Ingestion Framework
Configure Python and test the Ingestion Framework
Prerequisites
The Ingestion Framework is a Python module that wraps the OpenMetadata API and builds workflows and utilities on top of it. Therefore, you need to make sure that you have the complete OpenMetadata stack running: MySQL + ElasticSearch + OpenMetadata Server.
To do so, you can either build and run the OpenMetadata Server locally as well, or use the metadata CLI to spin up the Docker containers as explained in the Tooling Status section of this page.
Python Setup
We recommend using pyenv to properly install and manage different Python versions in your system. Note that OpenMetadata requires Python version +3.7. This doc might be helpful to set up the environment virtualization.
Using an M1 Mac? To ensure compatibility with the different dependencies, use Python version 3.9.8 or higher. Note that the code has not been tested with Python 3.10 due to some libraries not supporting that already.
For the instructions below, there are a couple of commands you'll need to run first to prepare your environment:
Generated Sources
The backbone of OpenMetadata is the series of JSON schemas defining the Entities and their properties.
All different parts of the code rely on those definitions. The first step to start developing new connectors is to properly set up your local environment to interact with the Entities.
In the Ingestion Framework, this process is handled with datamodel-code-generator, which is able to read JSON schemas and automatically prepare pydantic models representing the input definitions. Please, make sure to run make generate from the project root to fill the ingestion/src/metadata/generated directory with the required models.
Once you have generated the sources, you should be able to run the tests and the metadata CLI. You can test your setup by running make coverage and see if you get any errors.
Quality tools
When working on the Ingestion Framework, you might want to take into consideration the following style-check tooling:
- pylint is a Static Code Analysis tool to catch errors, align coding standards and help us follow conventions and apply improvements.
- black can be used to both autoformat the code and validate that the codebase is compliant.
- isort helps us not lose time trying to find the proper combination of importing from
stdlib, requirements, project files…
The main goal is to ensure standardized formatting throughout the codebase.
When developing, you can run these tools with make recipes: make lint, make black and make isort. Note that we are excluding the generated sources from the JSON Schema standards.
If you want to take this one step further and make sure that you are not committing any malformed changes, you can use pre-commit hooks. This is a powerful tool that allows us to run specific validations at commit time. If those validations fail, the commit won't proceed. The interesting point is that the tools are going to fix your code for you, so you can freely try to commit again!
You can install our hooks via make precommit_install.
Tooling Status
We are currently using:
pylint&blackin the CI validations, so make sure to review your PRs for any warnings you generated.black&isortin the pre-commit hooks.
If you're just developing on the Ingestion side, you might not need to run the server from Intellij and the simple docker approach might be good enough. In these scenarios, you can:
At least once, build the images fully:
Which will package the server code. Afterward, if the modifications only impact the ingestion code and need to refresh the ingestion container, you can run:
Running tests
To run the tests, you'll need to install some packages via make install_test.
You can validate the environment by running make coverage from the root directory. Note that from some tests, having the OpenMetadata server instance up is required as they interact with the API.
Debugging locally
Running the ingestion workflows from PyCharm and debugging them is quite useful. In order to do so, follow these steps:
First, make sure that your Python environment is pointing to the local env you are using to develop.
Then, you can prepare Run Configurations to execute the ingestion as you would run the CLI:
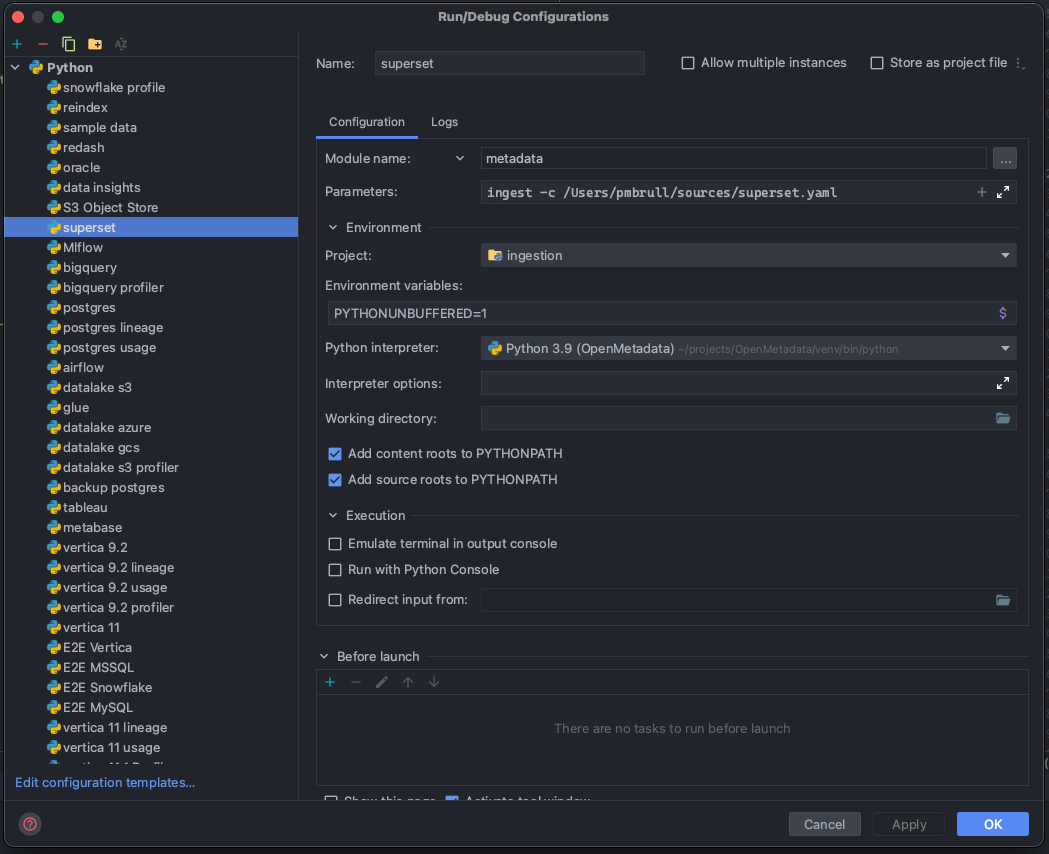
Note that in the example we are preparing a configuration to run and test Superset. In order to understand how to run ingestions via the CLI, you can refer to each connector's docs.
The important part is that we are not running a script, but rather a module: metadata. Based on this, we can work as we would usually do with the CLI for any ingestion, profiler, or test workflow.
With these settings, you can start adding breakpoints in PyCharm and debug the workflows!
Troubleshooting
If you try to install the kafka plugin pip install "./ingestion[kafka]" on mac you might see the following error:
This can be solved by running brew install librdkafka. If the error still persists you might need to update the following environment variables:
More information on the later in this thread.