Lineage Ingestion
A large subset of connectors distributed with OpenMetadata include support for lineage ingestion. Lineage ingestion processes queries to determine upstream and downstream entities for data assets. Lineage is published to the OpenMetadata catalog when metadata is ingested.
Using the OpenMetadata user interface and API, you may trace the path of data across Tables, Pipelines, and Dashboards.
Lineage ingestion is specific to the type of the Entity that we are processing. We are going to explain the ingestion process for the supported services.
The team is continuously working to increase the lineage coverage of the available services. Do not hesitate to reach out if you have any questions, issues or requests!
Database Services
Here we have 3 lineage sources, divided in different workflows, but mostly built around a Query Parser.
View Lineage
During the Metadata Ingestion workflow we differentiate if a Table is a View. For those sources where we can obtain the query that generates the View (e.g., Snowflake allows us to pick up the View query from the DDL).
After all Tables have been ingested in the workflow, it's time to parse all the queries generating Views. During the query parsing, we will obtain the source and target tables, search if the Tables exist in OpenMetadata, and finally create the lineage relationship between the involved Entities.
Let's go over this process with an example. Suppose have the following DDL:
From this query we will extract the following information:
1. There are two source tables, represented by the string schema.table_a as another_schema.table_b 2. There is a target table schema.my_view.
In this case we suppose that the database connection requires us to write the table names as <schema>.<table>. However, there are other possible options. Sometimes we can find just <table> in a query, or even <database>.<schema>.<table>.
The point here is that we have limited information that we can use to identify the Table Entity that represents the table written down in SQL. To close this gap, we run a query against ElasticSearch using the Table FQN.
Once we have identified all the ingredients in OpenMetadata as Entities, we can run the Lineage API to add the relationship between the nodes.
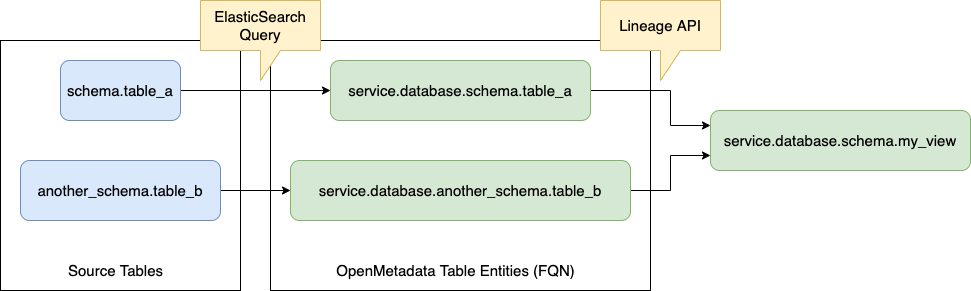
What we just described is the core process of identifying and ingesting lineage, and it will be reused (or partially reused) for the rest of the options as well.
dbt
When configuring an Ingestion adding dbt information we can parse the nodes on the Manifest JSON to get the data model lineage. Here we don't need to parse a query to obtain the source and target elements, but we still rely on querying ElasticSearch to identify the graph nodes as OpenMetadata Entities.
Note that if a Model is not materialized, its data won't be ingested.
How to run?
The main difference here is between those sources that provide internal access to query logs and those that do not. For services such as BigQuery, Snowflake etc.
There are specific workflows (Usage & Lineage) that will use the query log information. An alternative for sources not listed here is to run the workflow by providing the Query Logs that you have previously exported and then running the workflow.
Checkout the documentation of the connector you are using to know if it supports lineage & usage.
Process
That being said, this process is the same as the one shown in the View Lineage above. By obtaining a set of queries to parse, we will obtain the source and target information, use ElasticSearch to identify the Entities in OpenMetadata and then send the lineage to the API.
When running any query from within OpenMetadata we add an information comment to the query text
Note that queries with this text as well as the ones containing headers from dbt (which follow a similar structure), will be filtered out when building the query log internally.
Troubleshooting
Make sure that the tables that you are trying to add lineage for are present in OpenMetadata (and their upstream/downstream as well). You might also need to validate if the query logs are available in the tables for each service.
You can check the queries being used here:
By default, we apply a result limit of 1000 records. You might also need to increase that for databases with big volumes of queries.
Dashboard Services
When configuring the Ingestion Workflow for Dashboard Services you can select which Database Services are hosting the data feeding the Dashboards and Charts.
When ingesting the Dashboards metadata, the workflow will pick up the origin tables (or database, in the case of PowerBI), and prepare the lineage information.
Dashboard Lineage
Pipeline Services
The supported services here are Airflow, Fivetran, Dagster and Airbyte.
All of them ingest the lineage information out of the box. The only special case is Airflow, where one needs to setup inlets and outlets. You can find more information about it here.
Manual Lineage
Sometimes there is information that is shared among people but not present in the sources. To enable capturing all the possible knowledge, you can also add lineage manually with our UI editor.