Storage Services
This is the supported list of connectors for Storage Services:
If you have a request for a new connector, don't hesitate to reach out in Slack or open a feature request in our GitHub repo.
Configuring the Ingestion
In any other connector, extracting metadata happens automatically. We have different ways to understand the information in the sources and send that to OpenMetadata. However, what happens with generic sources such as S3 buckets, or ADLS containers?
In these systems we can have different types of information:
- Unstructured data, such as images or videos,
- Structured data in single and independent files (which can also be ingested with the S3 Data Lake connector)
- Structured data in partitioned files, e.g.,
my_table/year=2022/...parquet,my_table/year=2023/...parquet, etc.
The Storage Connector will help you bring in Structured data in partitioned files.
Then the question is, how do we know which data in each Container is relevant and which structure does it follow? In order to optimize ingestion costs and make sure we are only bringing in useful metadata, the Storage Services ingestion process follow this approach:
- We list the top-level containers (e.g., S3 buckets), and bring generic insights, such as size and number of objects.
- If there is an
openmetadata.jsonmanifest file present in the bucket root, we will ingest the informed paths as children of the top-level container. Let's see how that works.
Note that the current implementation brings each entry in the openmetadata.json as a child container of the top-level container. Even if your data path is s3://bucket/my/deep/table, we will bring bucket as the top-level container and my/deep/table as its child.
We are flattening this structure to simplify the navigation.
OpenMetadata Manifest
Our manifest file is defined as a JSON Schema, and can look like this:
Entries: We need to add a list of entries. Each inner JSON structure will be ingested as a child container of the top-level one. In this case, we will be ingesting 4 children.
Simple Container: The simplest container we can have would be structured, but without partitions. Note that we still need to bring information about:
- dataPath: Where we can find the data. This should be a path relative to the top-level container.
- structureFormat: What is the format of the data we are going to find. This information will be used to read the data.
- separator: Optionally, for delimiter-separated formats such as CSV, you can specify the separator to use when reading the file. If you don't, we will use
,for CSV and/tfor TSV files.
After ingesting this container, we will bring in the schema of the data in the dataPath.
Partitioned Container: We can ingest partitioned data without bringing in any further details.
By informing the isPartitioned field as true, we'll flag the container as Partitioned. We will be reading the source files schemas', but won't add any other information.
Single-Partition Container: We can bring partition information by specifying the partitionColumns. Their definition is based on the JSON Schema definition for table columns. The minimum required information is the name and dataType.
When passing partitionColumns, these values will be added to the schema, on top of the inferred information from the files.
Multiple-Partition Container: We can add multiple columns as partitions.
Note how in the example we even bring our custom displayName for the column dataTypeDisplay for its type.
Again, this information will be added on top of the inferred schema from the data files.
Automated Container Ingestion: Registering all the data paths one by one can be a time consuming job, to make the automated structure container ingestion you can provide the depth at which all the data is available.
Let us understand this with the example, suppose following is the file hierarchy within my bucket.
all my tables folders which contains the actual data are available at depth 3, hence when you specify the depth: 3 in manifest entry all following path will get registered as container in OpenMetadata with this single entry
saving efforts to add 4 individual entries compared to 1
Unstructured Container: OpenMetadata supports ingesting unstructured files like images, pdf's etc. We support fetching the file names, size and tags associates to such files.
In case you want to ingest a single unstructured file, then just specifying the full path of the unstructured file in datapath would be enough for ingestion.
In case you want to ingest all unstructured files with a specific extension for example pdf & png then you can provide the folder name containing such files in dataPath and list of extensions in the unstructuredFormats field.
In case you want to ingest all unstructured files with irrespective of their file type or extension then you can provide the folder name containing such files in dataPath and ["*"] in the unstructuredFormats field.
Global Manifest
You can also manage a single manifest file to centralize the ingestion process for any container, named openmetadata_storage_manifest.json. For example:
In that case, you will need to add a containerName entry to the structure above. For example:
The fields shown above (dataPath, structureFormat, isPartitioned, etc.) are still valid.
Container Name: Since we are using a single manifest for all your containers, the field containerName will help us identify which container (or Bucket in S3, etc.), contains the presented information.
You can also keep local manifests openmetadata.json in each container, but if possible, we will always try to pick up the global manifest during the ingestion.
Example
Let's show an example on how the data process and metadata look like. We will work with S3, using a global manifest, and two buckets.
S3 Data
In S3 we have:
- We have a bucket
om-glue-testwhere ouropenmetadata_storage_manifest.jsonglobal manifest lives. - We have another bucket
collate-demo-storagewhere we want to ingest the metadata of 5 partitioned containers with different formats- The
cities_multiple_simplecontainer has a time partition (formatting just a date) and aStatepartition. - The
cities_multiplecontainer has aYearand aStatepartition. - The
citiescontainer is only partitioned byState. - The
transactions_separatorcontainer contains multiple CSV files separated by;. - The
transactionscontainer contains multiple CSV files separated by,.
- The
The ingestion process will pick up a random sample of files from the directories (or subdirectories).
Global Manifest
Our global manifest looks like follows:
We are specifying:
- Where to find the data for each container we want to ingest via the
dataPath, - The
format, - Indication if the data has sub partitions or not (e.g.,
StateorYear), - The
containerName, so that the process knows in which S3 bucket to look for this data.
Source Config
In order to prepare the ingestion, we will:
- Set the
sourceConfigto include only the containers we are interested in. - Set the
storageMetadataConfigSourcepointing to the global manifest stored in S3, specifying the container name asom-glue-test.
You can run this same process from the UI, or directly with the metadata CLI via metadata ingest -c <path to yaml>.
Checking the results
Once the ingestion process runs, we'll see the following metadata:
First, the service we called s3-demo, which has the two buckets we included in the filter.
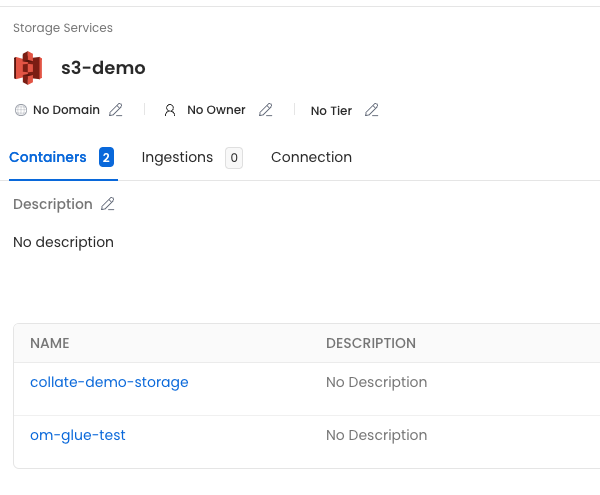
Then, if we click on the collate-demo-storage container, we'll see all the children defined in the manifest.
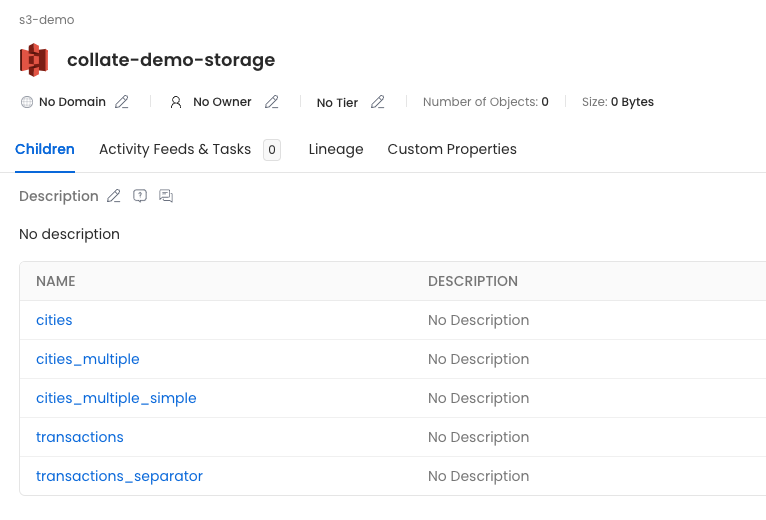
- cities: Will show the columns extracted from the sampled parquet files, since there is no partition columns specified.
- cities_multiple: Will have the parquet columns and the
YearandStatecolumns indicated in the partitions. - cities_multiple_simple: Will have the parquet columns and the
Statecolumn indicated in the partition. - transactions and transactions_separator: Will have the CSV columns.