Custom Connectors
Each of the services support providing a Custom Connector. It should be a Python class available in the Python environment running the ingestion process (e.g., EC2 instance, Airflow host, Docker Image...). It should also match specific constraints on the methods to implement and how to send the Entities to be created.
In this guide, we'll walk through a possible implementation. The example is based on a Database Service, but the process is the same for Pipelines, Dashboard or Messaging services.
This guide is based on a working example in the OpenMetadata Demos repository: link.
We'd recommend to go through the example to better understand how all the pieces should look like.
Watch OpenMetadata's Webinar on Custom Connectors
Steps to Set Up a Custom Connector
Step 1 - Prepare your Connector
A connector is a class that extends from from metadata.ingestion.api.steps import Source. It should implement all the required methods (docs).
In connector/my_awesome_connector.py you have a minimal example of it.
Note how te important method is the _iter. This is the generator function that will be iterated over to send all the Create Entity Requests to the Sink. Read more about the Workflow here.
Step 2 - Yield the Data
The Sink is expecting Create Entity Requests. To get familiar with the Python SDK and understand how to create the different Entities, a recommended read is the Python SDK docs.
We do not have docs and examples of all the supported Services. A way to get examples on how to create and fetch other types of Entities is to directly refer to the ometa integration tests.
Either & StackTraceError
When we yield the data, we are now wrapping the state of the execution being correct or not with an Either class:
This Either will have a left or right, and we will either return:
rightwith the correctCreateEntityRequestleftwith the exception that we want to track withStackTraceError.
For example:
Note that with the new structure, any errors are going to be properly logged at the end of the execution as:
Step 3 - Prepare the Package Installation
We'll need to package the code so that it can be shipped to the ingestion container and used there. In this demo you can find a simple setup.py that builds the connector module.
Step 4 - Prepare the Ingestion Image
If you want to use the connector from the UI, the Python environment running the ingestion process should contain the new code you just created. For example, if running via Docker, the openmetadata-ingestion image should be aware of your new package.
We will be running the demo against the OpenMetadata version 1.4.4, therefore, our Dockerfile looks like:
Step 5 - Run OpenMetadata with the Custom Ingestion Image
We have a Makefile prepared for you to run make run. This will get OpenMetadata up in Docker Compose using the custom Ingestion image.
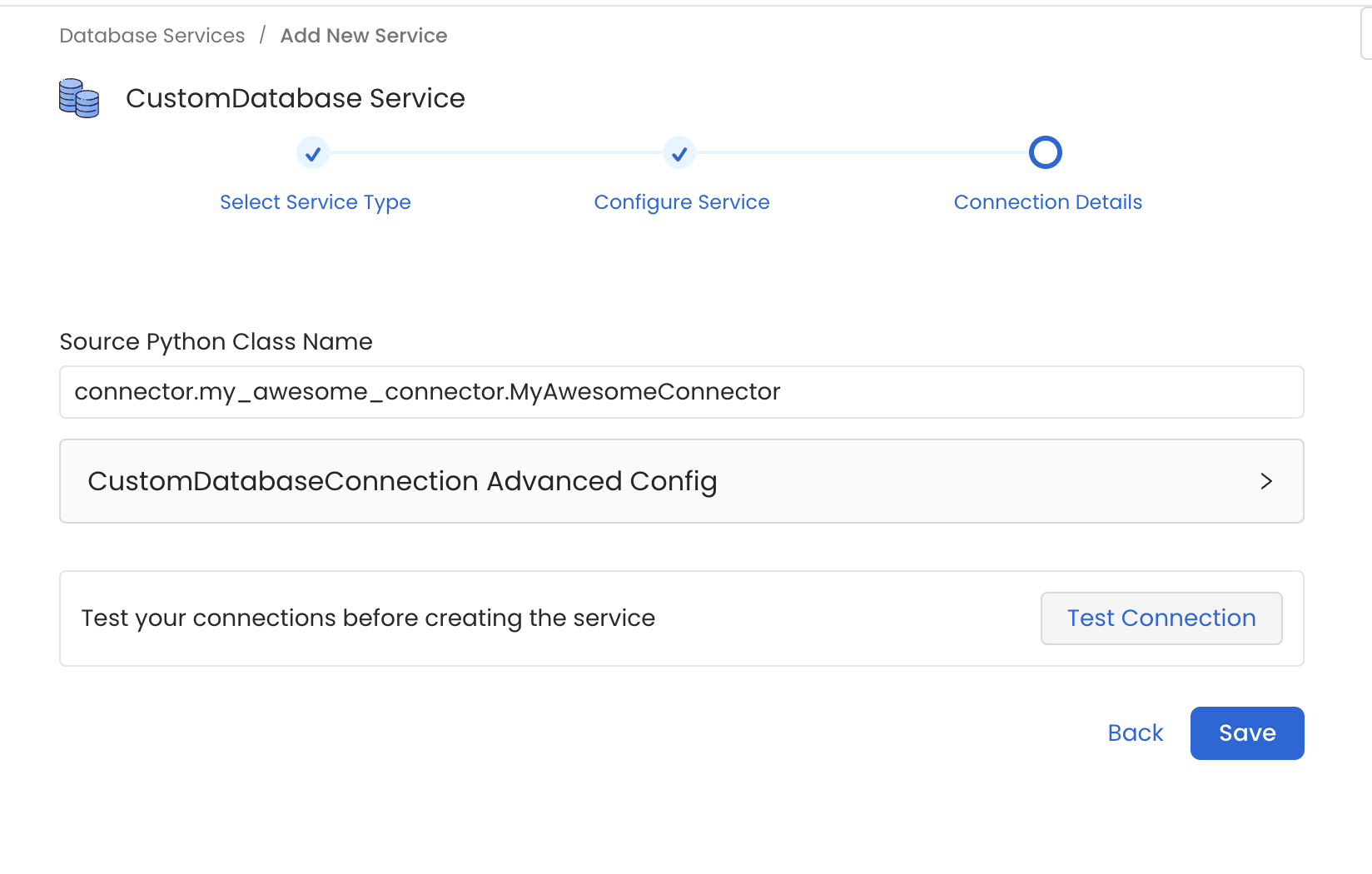
Step 6 - Configure the Connector
In the example we prepared a Database Connector. Thus, go to Database Services > Add New Service > Custom and set the Source Python Class Name as connector.my_awesome_connector.MyAwesomeConnector.
Note how we are specifying the full module name so that the Ingestion Framework can import the Source class.