KafkaConnect
PRODFeature List
✓ Pipelines
✓ Pipeline Status
✓ Lineage
✓ Usage
✕ Owners
✕ Tags
Requirements
KafkaConnect Versions
OpenMetadata is integrated with kafkaconnect up to version 3.6.1 and will continue to work for future kafkaconnect versions. The ingestion framework uses kafkaconnect python client to connect to the kafkaconnect instance and perform the API callsMetadata Ingestion
Visit the Services Page
Click `Settings` in the side navigation bar and then `Services`. The first step is to ingest the metadata from your sources. To do that, you first need to create a Service connection first. This Service will be the bridge between OpenMetadata and your source system. Once a Service is created, it can be used to configure your ingestion workflows.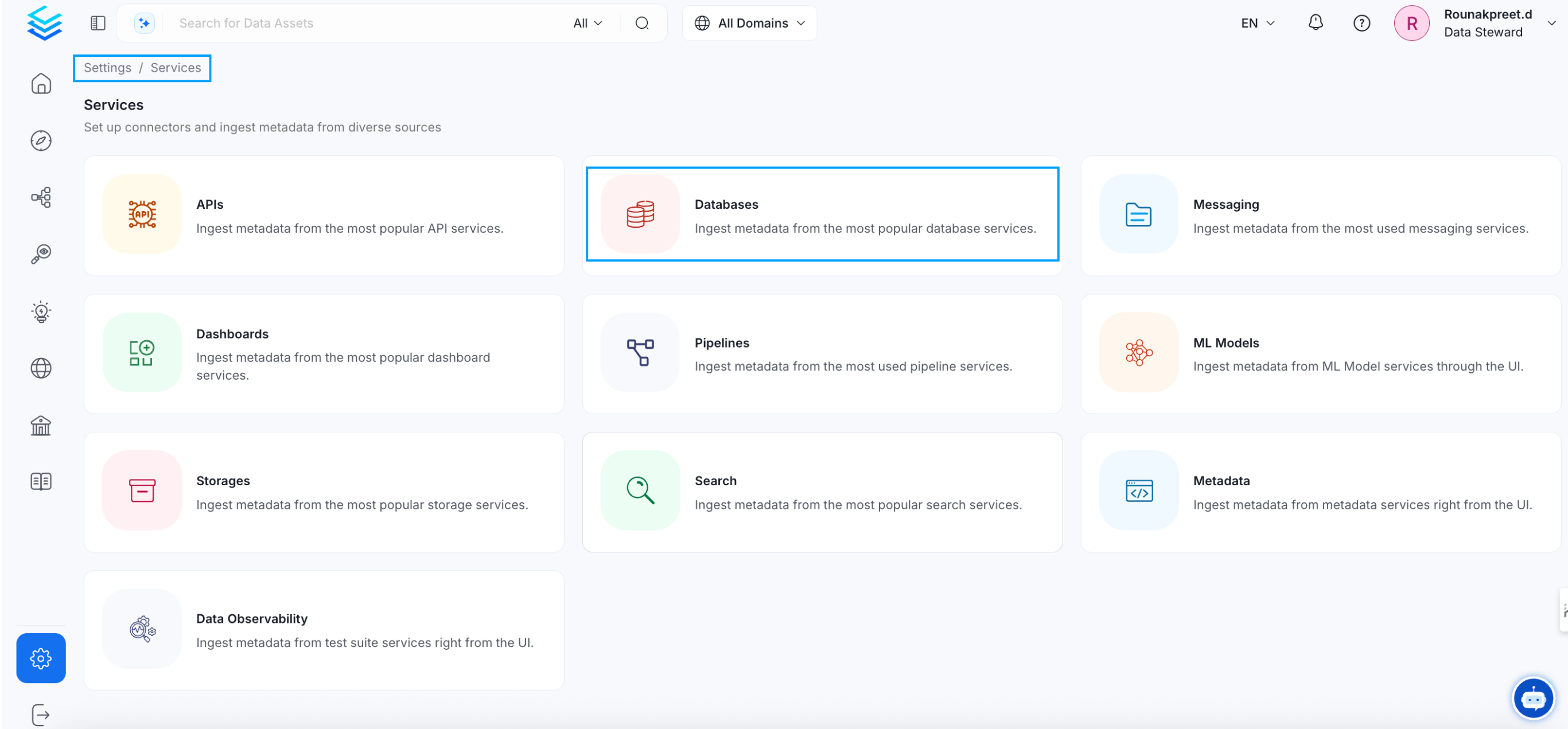

Name and Describe your Service
Provide a name and description for your Service.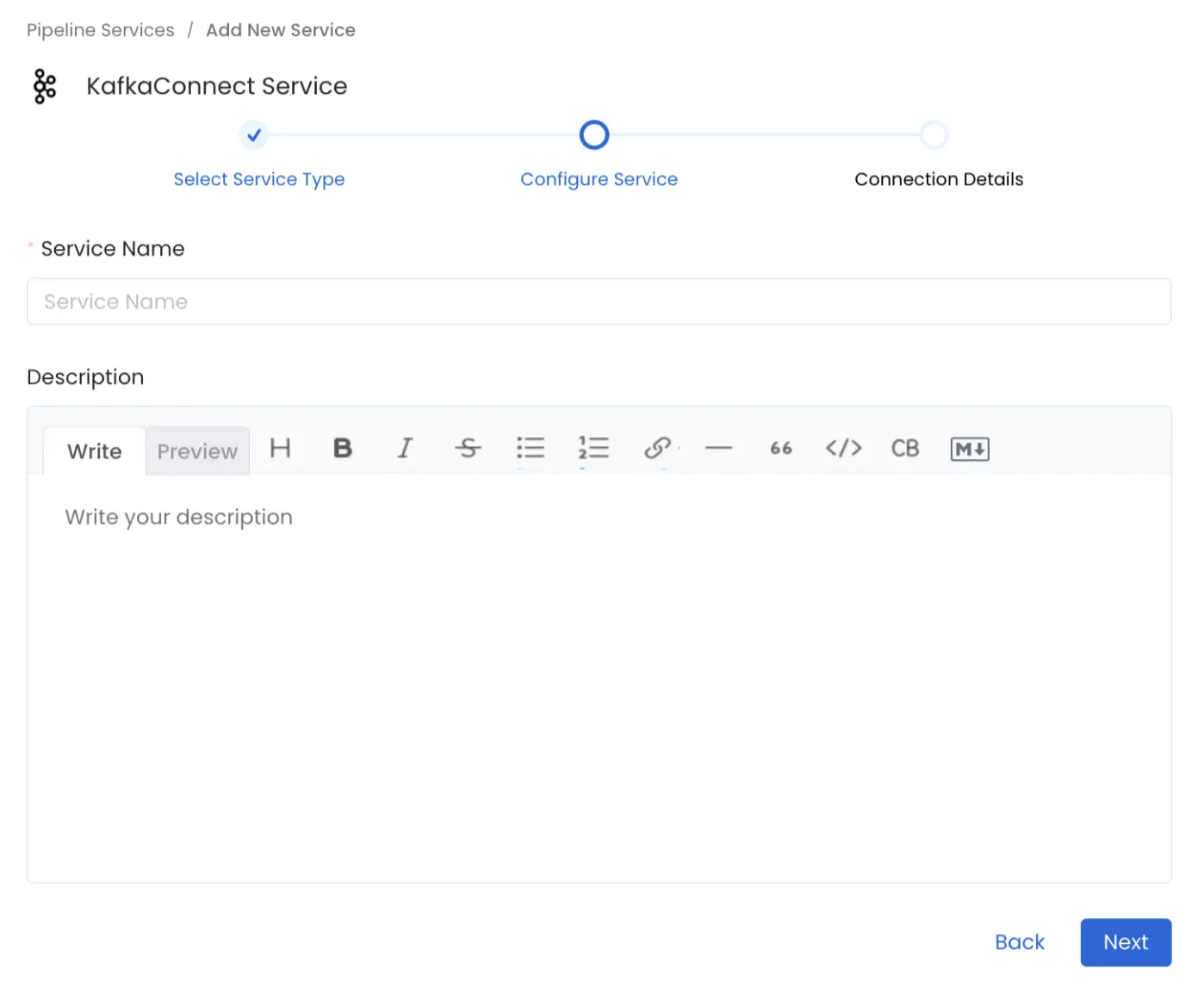
Service Name
OpenMetadata uniquely identifies Services by their **Service Name**. Provide a name that distinguishes your deployment from other Services, including the other KafkaConnect Services that you might be ingesting metadata from. Note that when the name is set, it cannot be changed.
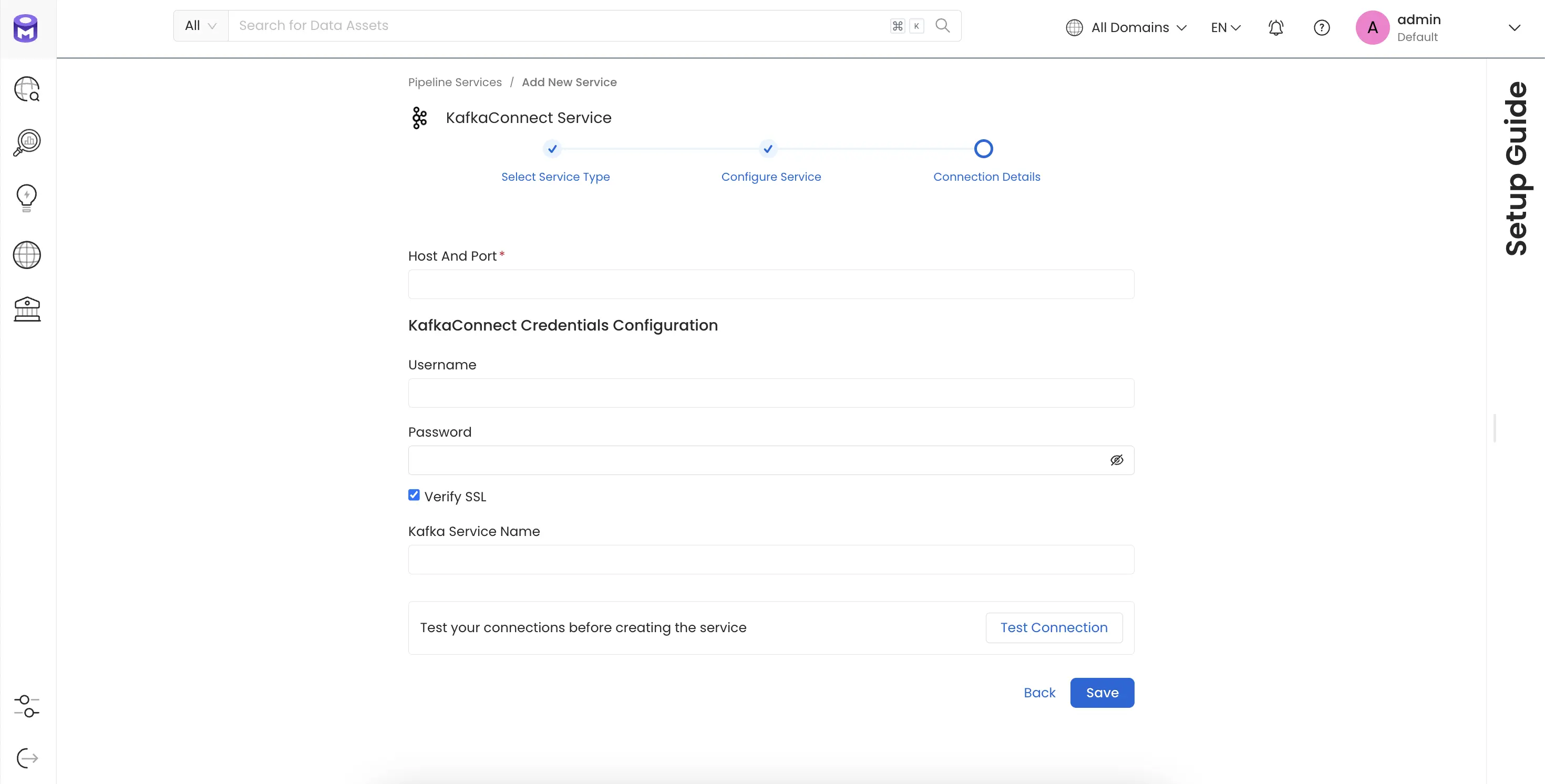
Connection Details
Connection Details
- Host and Port: The hostname or IP address of the Kafka Connect worker with the REST API enabled eg.
https://localhost:8083orhttps://127.0.0.1:8083orhttps://<yourkafkaconnectresthostnamehere> - Kafka Connect Config: OpenMetadata supports username/password.
- Basic Authentication
- Username: Username to connect to Kafka Connect. This user should be able to send request to the Kafka Connect API and access the Rest API GET endpoints.
- Password: Password to connect to Kafka Connect.
- Basic Authentication
- verifySSL : Whether SSL verification should be perform when authenticating.
- Kafka Service Name : The Service Name of the Ingested Kafka instance associated with this KafkaConnect instance.
Test the Connection
Once the credentials have been added, click on Test Connection and Save the changes.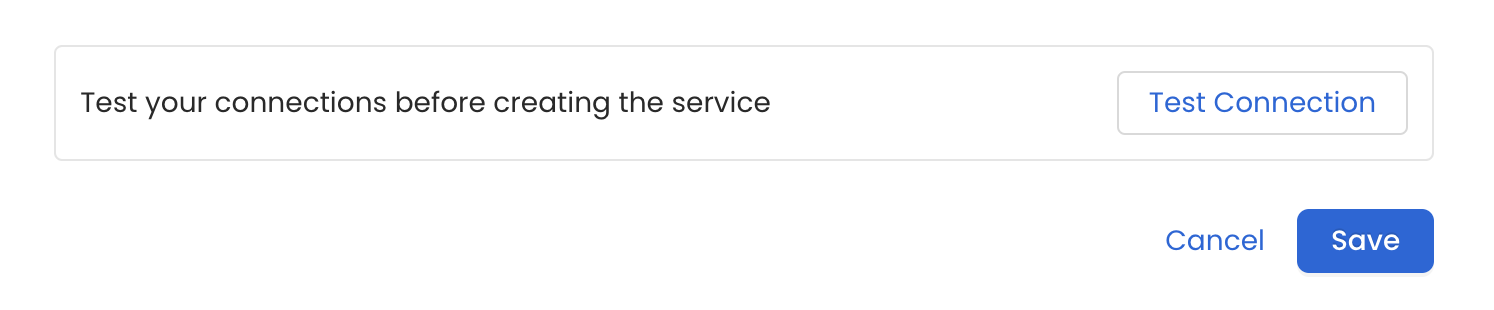

Configure Metadata Ingestion
In this step we will configure the metadata ingestion pipeline,
Please follow the instructions below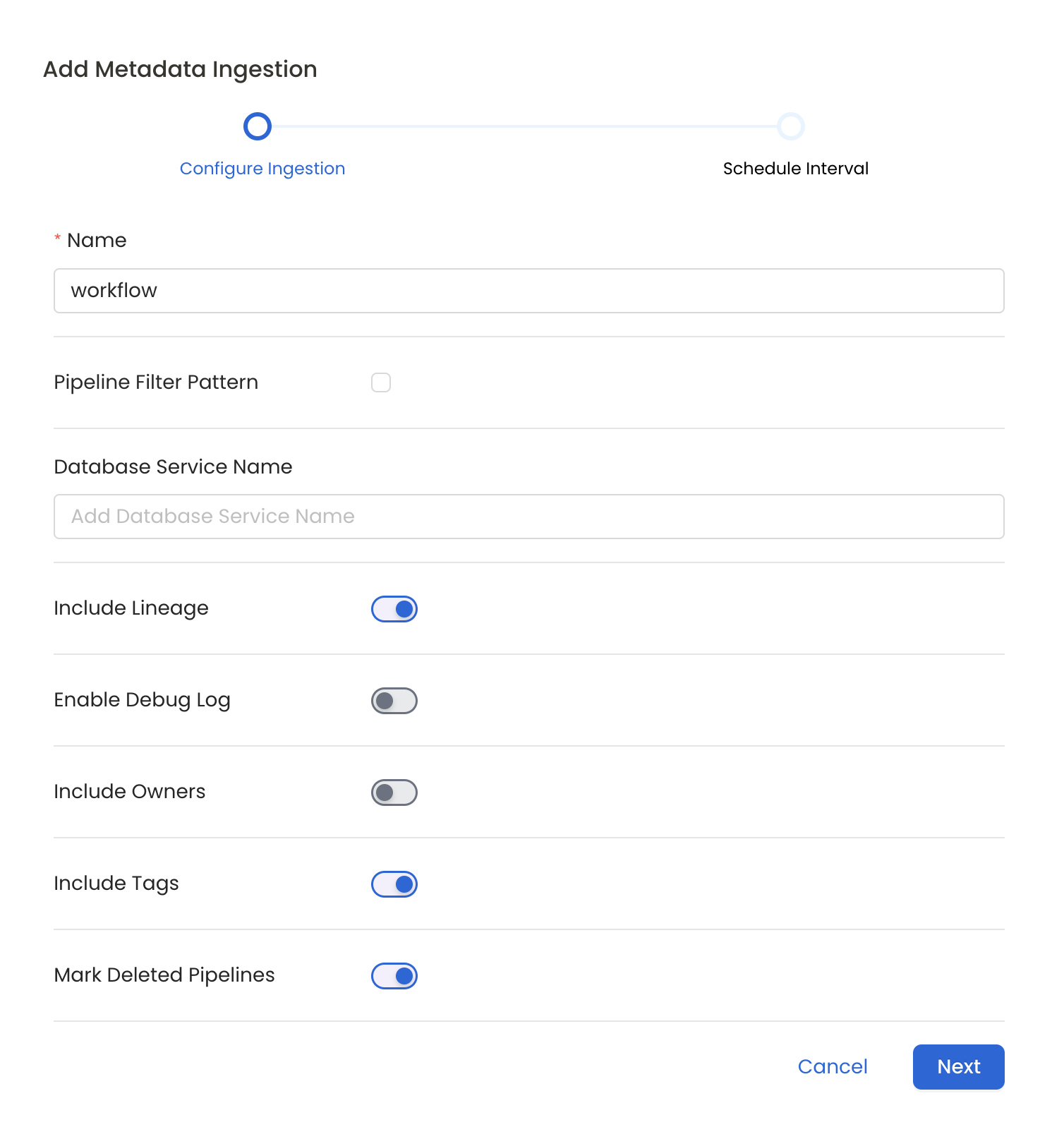

Metadata Ingestion Options
- Name: This field refers to the name of ingestion pipeline, you can customize the name or use the generated name.
- Pipeline Filter Pattern (Optional): Use to pipeline filter patterns to control whether or not to include pipeline as part of metadata ingestion.
- Include: Explicitly include pipeline by adding a list of comma-separated regular expressions to the Include field. OpenMetadata will include all pipeline with names matching one or more of the supplied regular expressions. All other schemas will be excluded.
- Exclude: Explicitly exclude pipeline by adding a list of comma-separated regular expressions to the Exclude field. OpenMetadata will exclude all pipeline with names matching one or more of the supplied regular expressions. All other schemas will be included.
- Include lineage (toggle): Set the Include lineage toggle to control whether to include lineage between pipelines and data sources as part of metadata ingestion.
- Enable Debug Log (toggle): Set the Enable Debug Log toggle to set the default log level to debug.
- Mark Deleted Pipelines (toggle): Set the Mark Deleted Pipelines toggle to flag pipelines as soft-deleted if they are not present anymore in the source system.
Schedule the Ingestion and Deploy
Scheduling can be set up at an hourly, daily, weekly, or manual cadence. The
timezone is in UTC. Select a Start Date to schedule for ingestion. It is
optional to add an End Date.Review your configuration settings. If they match what you intended,
click Deploy to create the service and schedule metadata ingestion.If something doesn’t look right, click the Back button to return to the
appropriate step and change the settings as needed.After configuring the workflow, you can click on Deploy to create the
pipeline.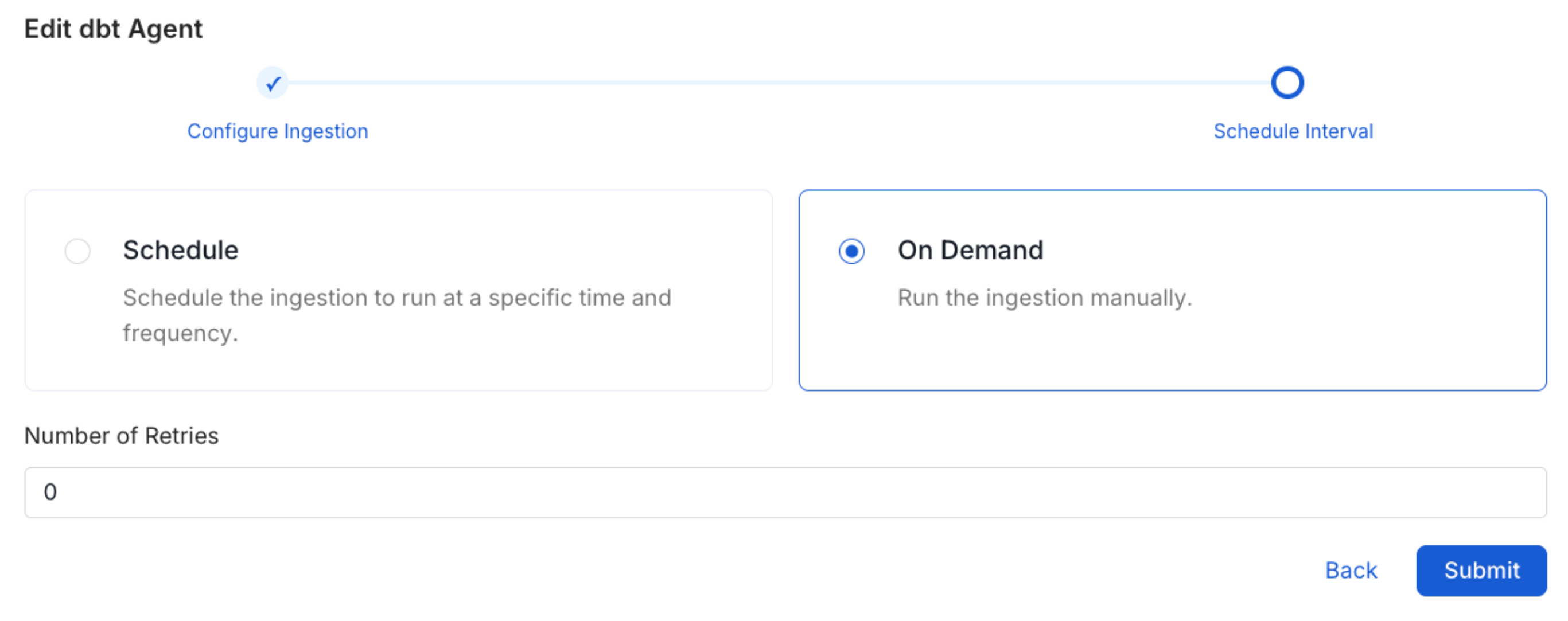

Debezium CDC Support
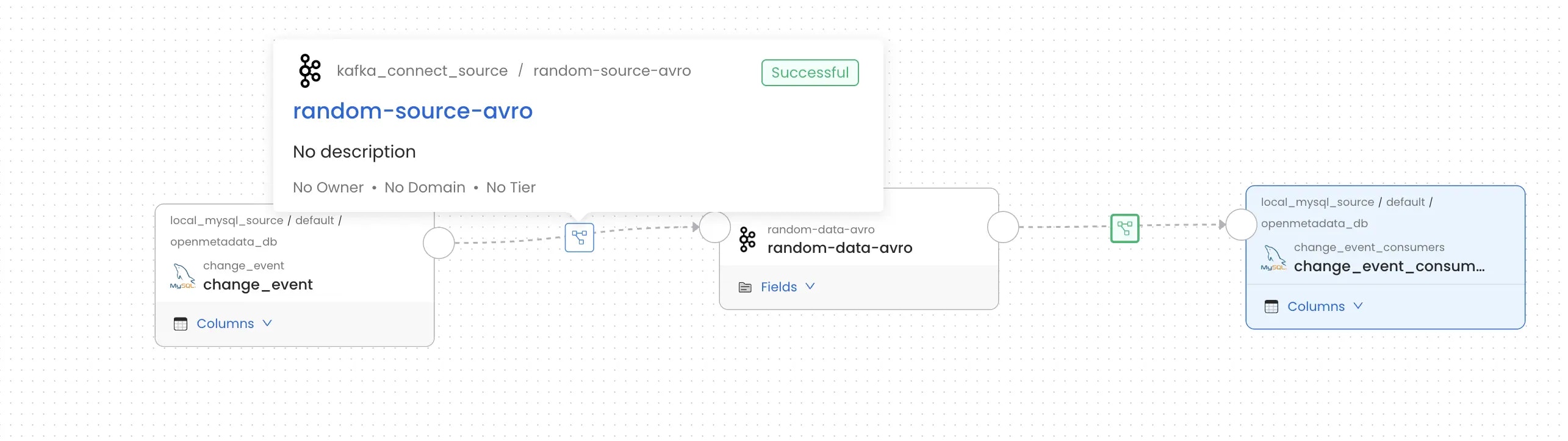
The KafkaConnect connector provides full support for Debezium CDC connectors with intelligent column extraction and accurate lineage tracking.What We Provide
When you ingest Debezium connectors, OpenMetadata automatically:- Detects CDC Envelope Structures - Identifies Debezium’s CDC format with
op,before, andafterfields - Extracts Real Table Columns - Parses actual database columns from the CDC payload instead of CDC envelope metadata
- Creates Accurate Column-Level Lineage - Maps lineage from source database tables → Kafka topics → target systems
Recognized Configuration Parameters
OpenMetadata recognizes the following Debezium configuration parameters for intelligent CDC detection:database.server.name- Server identifier (Debezium V1)topic.prefix- Topic prefix (Debezium V2)table.include.list- Tables to capture (e.g.,mydb.customers,mydb.orders)
Supported Connectors
Currently, the following source and sink connectors for Kafka Connect are supported for lineage tracking:- MySQL
- PostgreSQL
- MSSQL
- MongoDB
- Amazon S3 For these connectors, lineage information can be obtained provided they are configured with a source or sink and the corresponding metadata ingestion is enabled. Note: All supported database connectors listed above work seamlessly with Debezium CDC connectors for enhanced column-level lineage tracking. When using Debezium, OpenMetadata automatically detects the CDC envelope structure and extracts actual table columns for accurate lineage mapping.
Missing Lineage
If lineage information is not displayed for a Kafka Connect service, follow these steps to diagnose the issue.- Kafka Service Association: Make sure the Kafka service that the data is being ingested from is associated with this Kafka Connect service. Additionally, verify that the correct name is passed on in the Kafka Service Name field during configuration. This field helps establish the lineage between the Kafka service and the Kafka Connect flow.
- Source and Sink Configuration: Verify that the Kafka Connect connector associated with the service is configured with a source and/or sink database or storage system. Connectors without a defined source or sink cannot provide lineage data.
- Metadata Ingestion: Ensure that metadata for both the source and sink database/storage systems is ingested and passed to the lineage system. This typically involves configuring the relevant connectors to capture and transmit this information.