This page contains the setup guide and reference information for the GCS connector.
Configure and schedule GCS metadata workflows from the OpenMetadata UI:
Ingestion Deployment
To run the Ingestion via the UI you'll need to use the OpenMetadata Ingestion Container, which comes shipped with custom Airflow plugins to handle the workflow deployment. If you want to install it manually in an already existing Airflow host, you can follow this guide.
If you don't want to use the OpenMetadata Ingestion container to configure the workflows via the UI, then you can check the following docs to run the Ingestion Framework in any orchestrator externally.
Run Connectors from the OpenMetadata UI
Learn how to manage your deployment to run connectors from the UIRun the Connector Externally
Get the YAML to run the ingestion externallyExternal Schedulers
Get more information about running the Ingestion Framework ExternallyRequirements
We need the following permissions in GCP:
GCS Permissions
For all the buckets that we want to ingest, we need to provide the following:
storage.buckets.getstorage.buckets.liststorage.objects.getstorage.objects.list
OpenMetadata Manifest
In any other connector, extracting metadata happens automatically. In this case, we will be able to extract high-level metadata from buckets, but in order to understand their internal structure we need users to provide an openmetadata.json file at the bucket root.
Supported File Formats: [ "csv", "tsv", "avro", "parquet", "json", "json.gz", "json.zip" ]
You can learn more about this here. Keep reading for an example on the shape of the manifest file.
OpenMetadata Manifest
Our manifest file is defined as a JSON Schema, and can look like this:
Entries: We need to add a list of entries. Each inner JSON structure will be ingested as a child container of the top-level one. In this case, we will be ingesting 4 children.
Simple Container: The simplest container we can have would be structured, but without partitions. Note that we still need to bring information about:
- dataPath: Where we can find the data. This should be a path relative to the top-level container.
- structureFormat: What is the format of the data we are going to find. This information will be used to read the data.
- separator: Optionally, for delimiter-separated formats such as CSV, you can specify the separator to use when reading the file. If you don't, we will use
,for CSV and/tfor TSV files.
After ingesting this container, we will bring in the schema of the data in the dataPath.
Partitioned Container: We can ingest partitioned data without bringing in any further details.
By informing the isPartitioned field as true, we'll flag the container as Partitioned. We will be reading the source files schemas', but won't add any other information.
Single-Partition Container: We can bring partition information by specifying the partitionColumns. Their definition is based on the JSON Schema definition for table columns. The minimum required information is the name and dataType.
When passing partitionColumns, these values will be added to the schema, on top of the inferred information from the files.
Multiple-Partition Container: We can add multiple columns as partitions.
Note how in the example we even bring our custom displayName for the column dataTypeDisplay for its type.
Again, this information will be added on top of the inferred schema from the data files.
Global Manifest
You can also manage a single manifest file to centralize the ingestion process for any container, named openmetadata_storage_manifest.json. For example:
In that case, you will need to add a containerName entry to the structure above. For example:
The fields shown above (dataPath, structureFormat, isPartitioned, etc.) are still valid.
Container Name: Since we are using a single manifest for all your containers, the field containerName will help us identify which container (or Bucket in S3, etc.), contains the presented information.
You can also keep local manifests openmetadata.json in each container, but if possible, we will always try to pick up the global manifest during the ingestion.
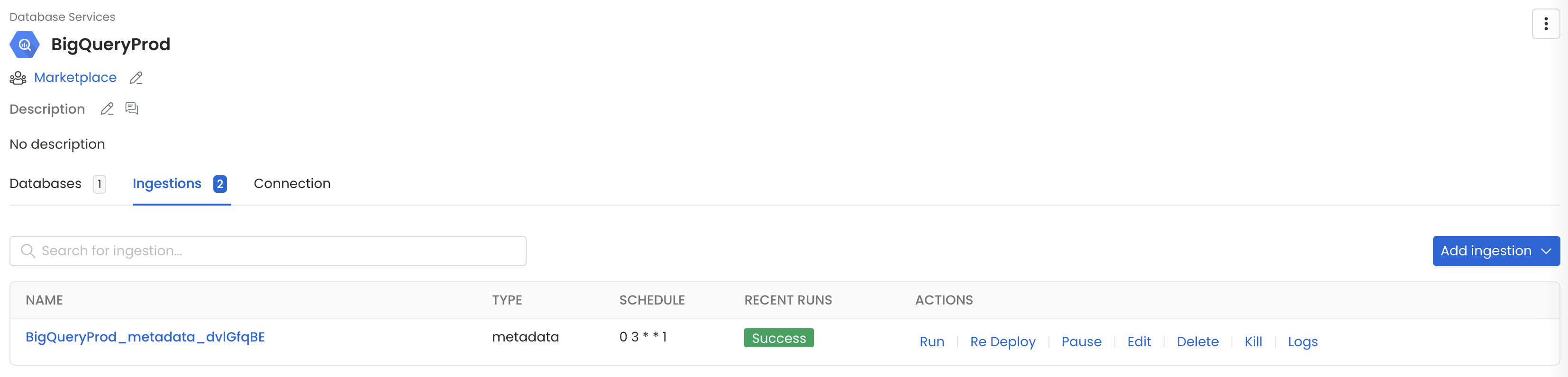
Metadata Ingestion
1. Visit the Services Page
The first step is ingesting the metadata from your sources. Under Settings, you will find a Services link an external source system to OpenMetadata. Once a service is created, it can be used to configure metadata, usage, and profiler workflows.
To visit the Services page, select Services from the Settings menu.
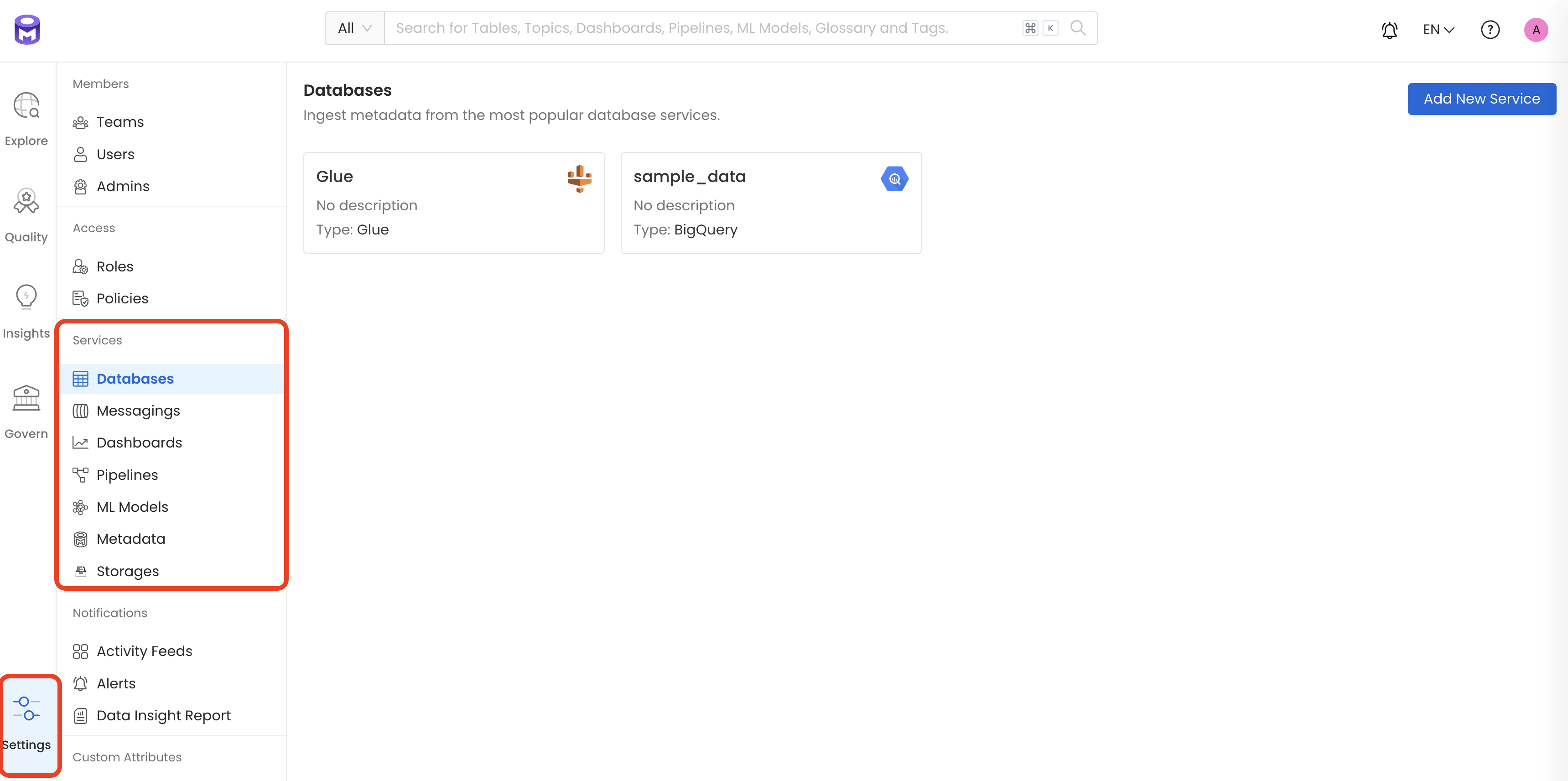
Find Dashboard option on left panel of the settings page
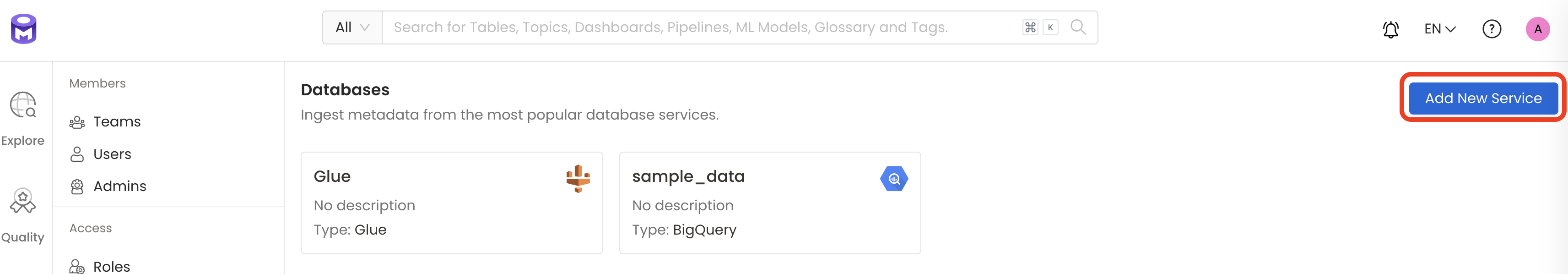
Add a new Service from the Storage Services page

Select your service from the list
5. Configure the Service Connection
In this step, we will configure the connection settings required for this connector. Please follow the instructions below to ensure that you've configured the connector to read from your GCS service as desired.

Configure the service connection by filling the form
6. Test the Connection
Once the credentials have been added, click on Test Connection and Save the changes.
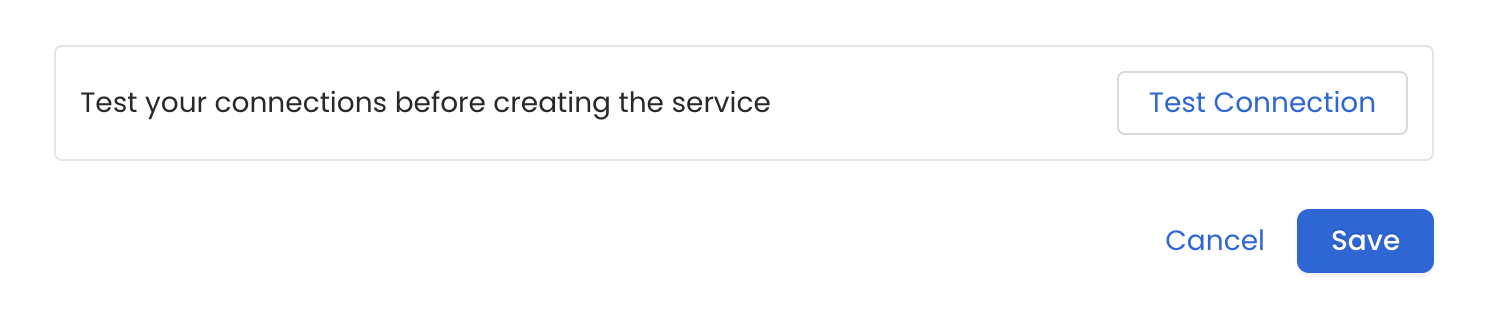
Test the connection and save the Service
7. Configure Metadata Ingestion
In this step we will configure the metadata ingestion pipeline, Please follow the instructions below
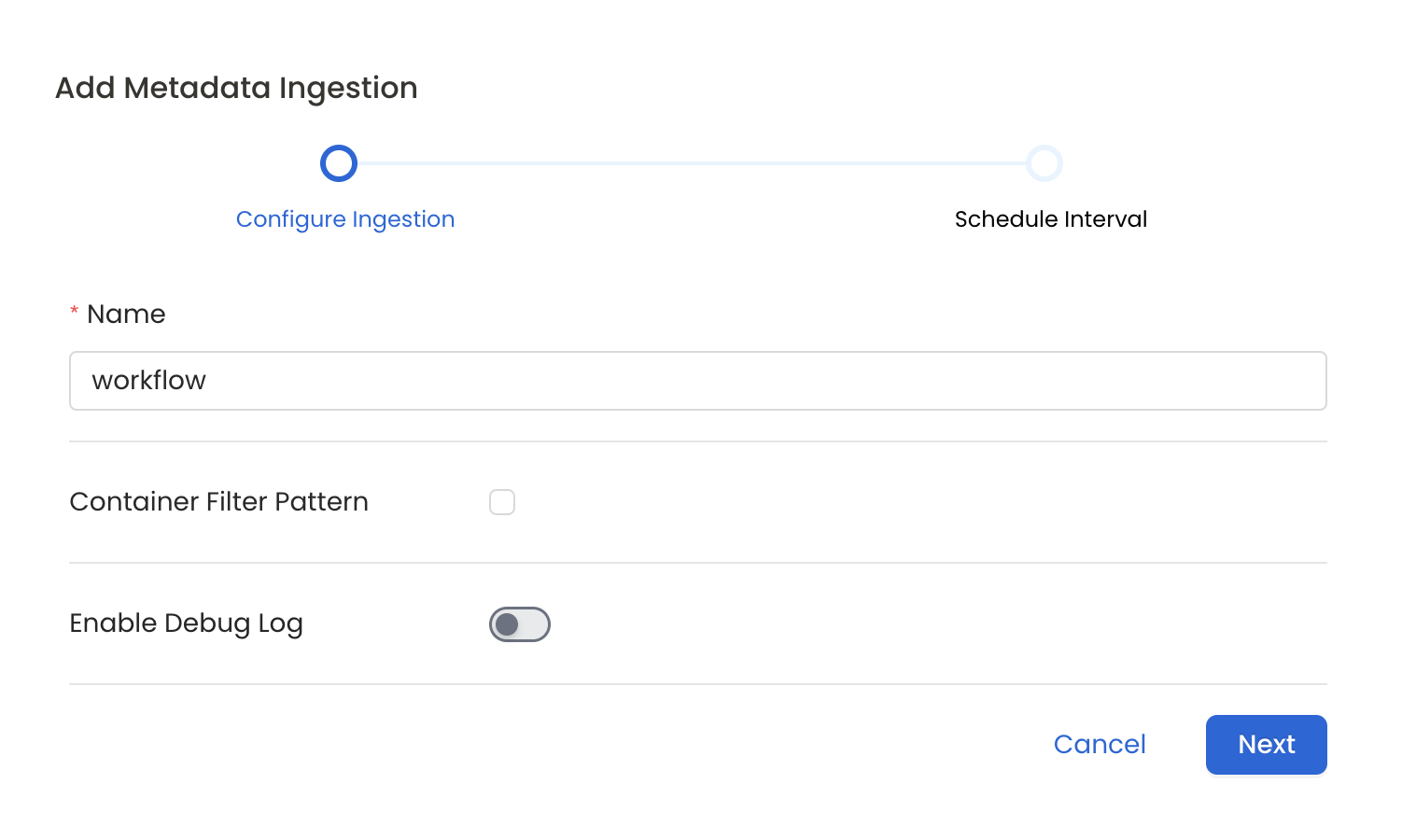
Configure Metadata Ingestion Page
Metadata Ingestion Options
- Name: This field refers to the name of ingestion pipeline, you can customize the name or use the generated name.
- Container Filter Pattern (Optional): To control whether to include a container as part of metadata ingestion.
- Include: Explicitly include containers by adding a list of comma-separated regular expressions to the Include field. OpenMetadata will include all containers with names matching one or more of the supplied regular expressions. All other containers will be excluded.
- Exclude: Explicitly exclude containers by adding a list of comma-separated regular expressions to the Exclude field. OpenMetadata will exclude all containers with names matching one or more of the supplied regular expressions. All other containers will be included.
- Enable Debug Log (toggle): Set the Enable Debug Log toggle to set the default log level to debug.
- Storage Metadata Config Source: Here you can specify the location of your global manifest
openmetadata_storage_manifest.jsonfile. It can be located in S3, a local path or HTTP.
8. Schedule the Ingestion and Deploy
Scheduling can be set up at an hourly, daily, weekly, or manual cadence. The timezone is in UTC. Select a Start Date to schedule for ingestion. It is optional to add an End Date.
Review your configuration settings. If they match what you intended, click Deploy to create the service and schedule metadata ingestion.
If something doesn't look right, click the Back button to return to the appropriate step and change the settings as needed.
After configuring the workflow, you can click on Deploy to create the pipeline.
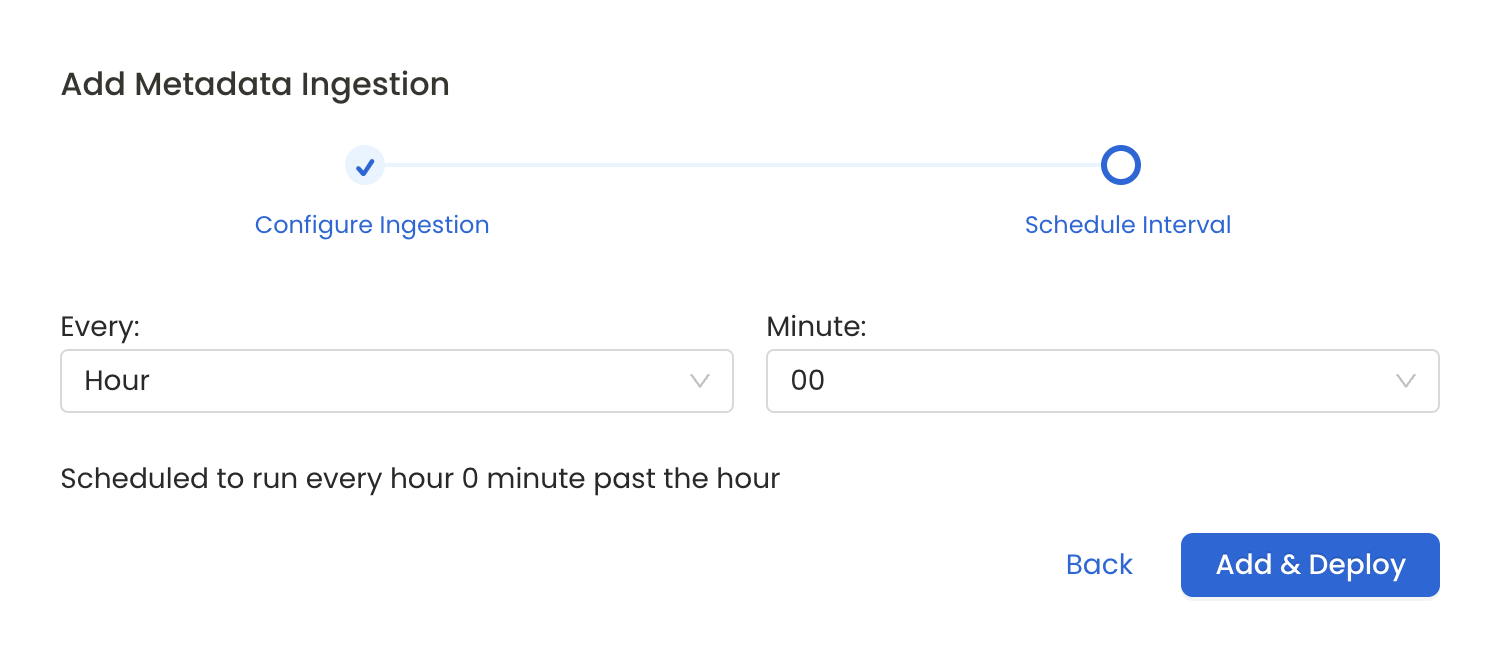
Schedule the Ingestion Pipeline and Deploy
Troubleshooting
Workflow Deployment Error
If there were any errors during the workflow deployment process, the Ingestion Pipeline Entity will still be created, but no workflow will be present in the Ingestion container.
- You can then Edit the Ingestion Pipeline and Deploy it again.
- From the Connection tab, you can also Edit the Service if needed.