How to Deploy a Lineage Workflow
Lineage data can be ingested from your data sources right from the OpenMetadata UI. Currently, the lineage workflow is supported for a limited set of connectors, like BigQuery, Snowflake, MSSQL, Redshift, Clickhouse, PostgreSQL, Databricks.
Tip: Trace the upstream and downstream dependencies with Lineage.
View Lineage from Metadata Ingestion
Once the metadata ingestion runs correctly, and we are able to explore the service Entities, we can add the view lineage information for the data assets. This will populate the Lineage tab in the data asset page. During the Metadata Ingestion workflow we differentiate if a Table is a View. For those sources, where we can obtain the query that generates the View, we bring in the view lineage along with the metadata. After all Tables have been ingested in the workflow, it's time to parse all the queries generating Views. During the query parsing, we will obtain the source and target tables, search if the Tables exist in OpenMetadata, and finally create the lineage relationship between the involved Entities.
If the database has views, then the view lineage would be generated automatically, along with the column-level lineage. In such a case, the table type is View as shown in the example below.
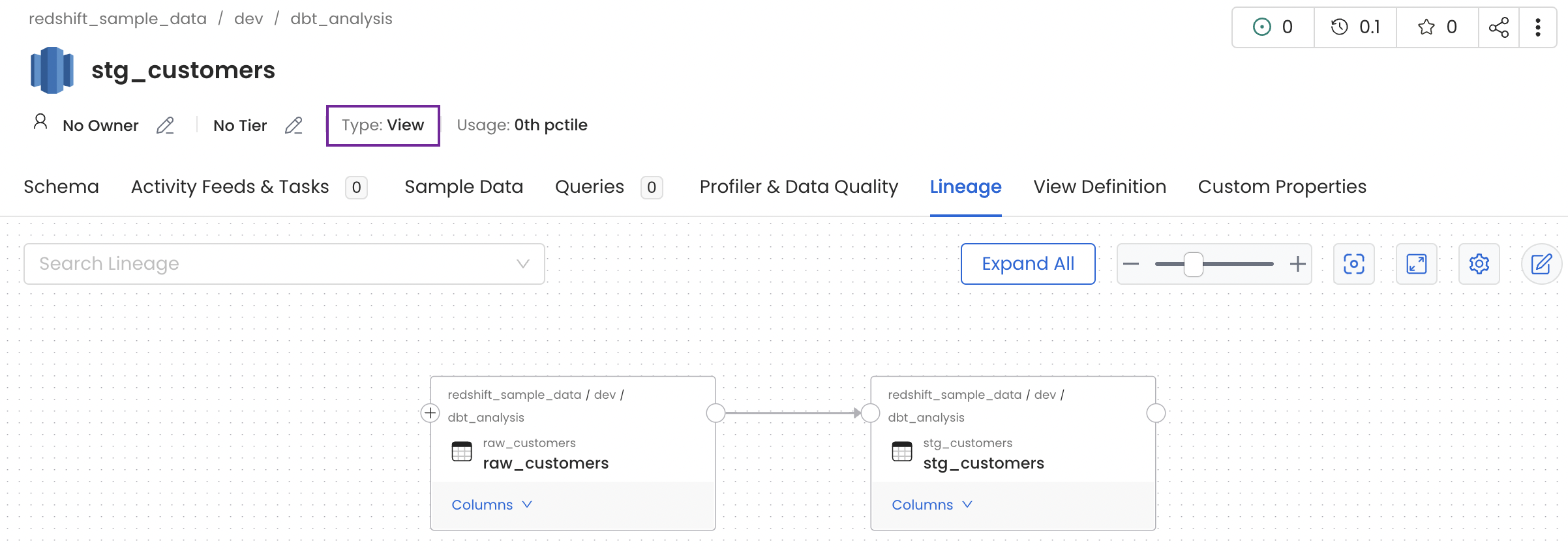
View Lineage through Metadata Ingestion
Lineage Agent from UI
Apart from the Metadata ingestion, we can create a workflow that will obtain the query log and table creation information from the underlying database and feed it to OpenMetadata. The Lineage Agent will be in charge of obtaining this data. The metadata ingestion will only bring in the View lineage queries, whereas the Lineage Agent workflow will be bring in all those queries that can be used to generate lineage information.
1. Add a Lineage Agent
Navigate to Settings >> Services >> Databases. Select the required service
Select a Service
Click on Databases
Select the Database
Go the the Agents tab. Click on Add Agent and select Add Lineage Agent.
Add a Lineage Agent
2. Configure the Lineage Agent
Here you can enter the Lineage Agent details:
Configure the Lineage Agent
Lineage Options
Query Log Duration: Specify the duration in days for which the profiler should capture lineage data from the query logs. For example, if you specify 2 as the value for the duration, the data profiler will capture lineage information for 2 days or 48 hours prior to when the ingestion workflow is run.
Parsing Timeout Limit: Specify the timeout limit for parsing the sql queries to perform the lineage analysis. This must be specified in seconds.
Result Limit: Set the limit for the query log results to be run at a time. This is the number of rows.
Filter Condition: We execute a query on query history table of the respective data source to perform the query analysis and extract the lineage and usage information. This field will be useful when you want to restrict some queries from being part of this analysis. In this field you can specify a sql condition that will be applied on the query history result set. You can check more about Usage Query Filtering here.
3. Schedule and Deploy
After clicking Next, you will be redirected to the Scheduling form. This will be the same as the Metadata Ingestion. Select your desired schedule and click on Deploy to find the lineage pipeline being added to the Service Ingestions.
Schedule and Deploy the Lineage Agent
Run Lineage Workflow Externally
Lineage
After running a Metadata Ingestion workflow, we can run Lineage workflow. While the serviceName will be the same to that was used in Metadata Ingestion, so the ingestion bot can get the serviceConnection details from the server.
1. Define the YAML Config
This is a sample config for BigQuery Lineage:
Source Configuration - Source Config
You can find all the definitions and types for the sourceConfig here.
queryLogDuration: Configuration to tune how far we want to look back in query logs to process lineage data in days.
parsingTimeoutLimit: Configuration to set the timeout for parsing the query in seconds.
filterCondition: Condition to filter the query history.
resultLimit: Configuration to set the limit for query logs.
queryLogFilePath: Configuration to set the file path for query logs.
databaseFilterPattern: Regex to only fetch databases that matches the pattern.
schemaFilterPattern: Regex to only fetch tables or databases that matches the pattern.
tableFilterPattern: Regex to only fetch tables or databases that matches the pattern.
overrideViewLineage: Set the 'Override View Lineage' toggle to control whether to override the existing view lineage.
processViewLineage: Set the 'Process View Lineage' toggle to control whether to process view lineage.
processQueryLineage: Set the 'Process Query Lineage' toggle to control whether to process query lineage.
processStoredProcedureLineage: Set the 'Process Stored ProcedureLog Lineage' toggle to control whether to process stored procedure lineage.
threads: Number of Threads to use in order to parallelize lineage ingestion.
- You can learn more about how to configure and run the Lineage Workflow to extract Lineage data from here
2. Run with the CLI
After saving the YAML config, we will run the command the same way we did for the metadata ingestion:
dbt Ingestion
We can also generate lineage through dbt ingestion. The dbt workflow can fetch queries that carry lineage information. For a dbt ingestion pipeline, the path to the Catalog and Manifest files must be specified. We also fetch the column level lineage through dbt.
You can learn more about lineage ingestion here.
Query Logs using CSV File
Lineage ingestion is supported for a few connectors as mentioned earlier. For the unsupported connectors, you can set up Lineage Workflows using Query Logs using a CSV file.
Manual Lineage
Lineage can also be added and edited manually in OpenMetadata. Refer for more information on adding lineage manually.
Explore the Lineage ViewExplore the rich lineage view in OpenMetadata.