BigQuery
PRODIn this section, we provide guides and references to use the BigQuery connector.
Configure and schedule BigQuery metadata and profiler workflows from the OpenMetadata UI:
- Requirements
- Metadata Ingestion
- Query Usage
- Data Profiler
- Data Quality
- Lineage
- dbt Integration
- Troubleshooting
Ingestion Deployment
To run the Ingestion via the UI you'll need to use the OpenMetadata Ingestion Container, which comes shipped with custom Airflow plugins to handle the workflow deployment. If you want to install it manually in an already existing Airflow host, you can follow this guide.
If you don't want to use the OpenMetadata Ingestion container to configure the workflows via the UI, then you can check the following docs to run the Ingestion Framework in any orchestrator externally.
Run Connectors from the OpenMetadata UI
Learn how to manage your deployment to run connectors from the UIRun the Connector Externally
Get the YAML to run the ingestion externallyExternal Schedulers
Get more information about running the Ingestion Framework ExternallyHow to Run the Connector Externally
To run the Ingestion via the UI you'll need to use the OpenMetadata Ingestion Container, which comes shipped with custom Airflow plugins to handle the workflow deployment.
If, instead, you want to manage your workflows externally on your preferred orchestrator, you can check the following docs to run the Ingestion Framework anywhere.
Requirements
You need to create an service account in order to ingest metadata from bigquery refer this guide on how to create service account.
Partitioned Tables
When profiling partitioned tables in BigQuery, OpenMetadata applies a default partition query duration of 1 day for time-based partitions. This conservative setting prevents excessive data scans but may result in no Sample Data or Column Profile Metrics if no data falls within the default window.
Resolution
You can adjust this behavior directly from the UI:
- Navigate to the table's detail page.
- Edit the profiler configuration.
- Update the
partitionQueryDurationunder Partition Config to a wider window (e.g., 30 days) as needed.
This change allows OpenMetadata to access a broader data range during profiling and sample data collection, resolving the issue for partitioned tables.
Data Catalog API Permissions
- Go to https://console.cloud.google.com/apis/library/datacatalog.googleapis.com
- Select the
GCP Project IDthat you want to enable theData Catalog APIon. - Click on
Enable APIwhich will enable the data catalog api on the respective project.
Access to the Google Data Catalog API is optional and only required if you want to retrieve policy tags from BigQuery. The BigQuery connector does not require this permission for general metadata ingestion.
GCP Permissions
To execute metadata extraction and usage workflow successfully the user or the service account should have enough access to fetch required data. Following table describes the minimum required permissions
| # | GCP Permission | Required For |
|---|---|---|
| 1 | bigquery.datasets.get | Metadata Ingestion |
| 2 | bigquery.tables.get | Metadata Ingestion |
| 3 | bigquery.tables.getData | Metadata Ingestion |
| 4 | bigquery.tables.list | Metadata Ingestion |
| 5 | resourcemanager.projects.get | Metadata Ingestion |
| 6 | bigquery.jobs.create | Metadata Ingestion |
| 7 | bigquery.jobs.listAll | Metadata Ingestion |
| 8 | bigquery.routines.get | Stored Procedure |
| 9 | bigquery.routines.list | Stored Procedure |
| 10 | datacatalog.taxonomies.get | Fetch Policy Tags |
| 11 | datacatalog.taxonomies.list | Fetch Policy Tags |
| 12 | bigquery.readsessions.create | Bigquery Usage & Lineage Workflow |
| 13 | bigquery.readsessions.getData | Bigquery Usage & Lineage Workflow |
| 14 | logging.operations.list | Incremental Metadata Ingestion |
If the user has External Tables, please attach relevant permissions needed for external tables, alongwith the above list of permissions.
If you are using BigQuery and have sharded tables, you might want to consider using partitioned tables instead. Partitioned tables allow you to efficiently query data by date or other criteria, without having to manage multiple tables. Partitioned tables also have lower storage and query costs than sharded tables. You can learn more about the benefits of partitioned tables here. If you want to convert your existing sharded tables to partitioned tables, you can follow the steps in this guide. This will help you simplify your data management and optimize your performance in BigQuery.
Metadata Ingestion
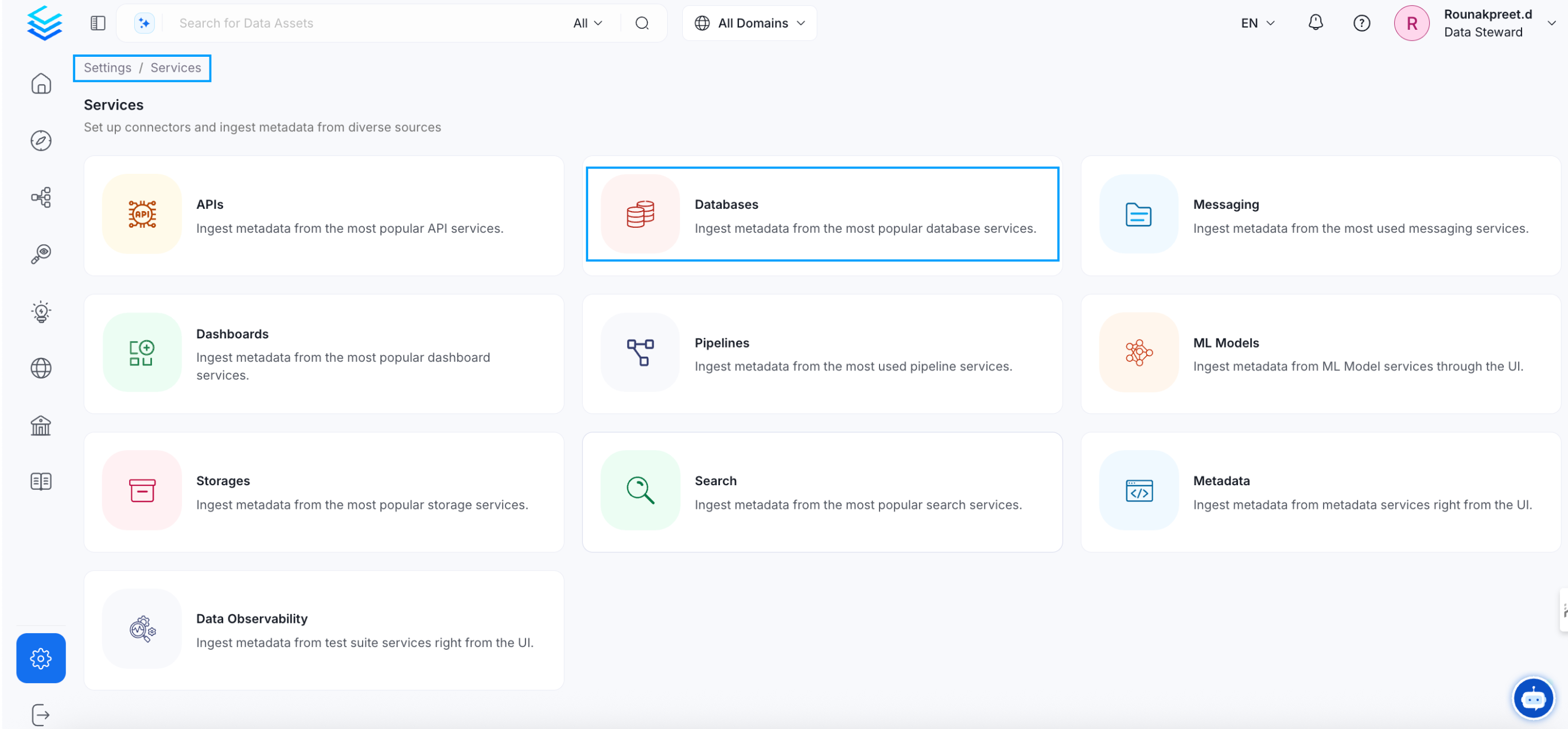
1. Visit the Services Page
Click Settings in the side navigation bar and then Services.
The first step is to ingest the metadata from your sources. To do that, you first need to create a Service connection first.
This Service will be the bridge between OpenMetadata and your source system.
Once a Service is created, it can be used to configure your ingestion workflows.
Select your Service Type and Add a New Service
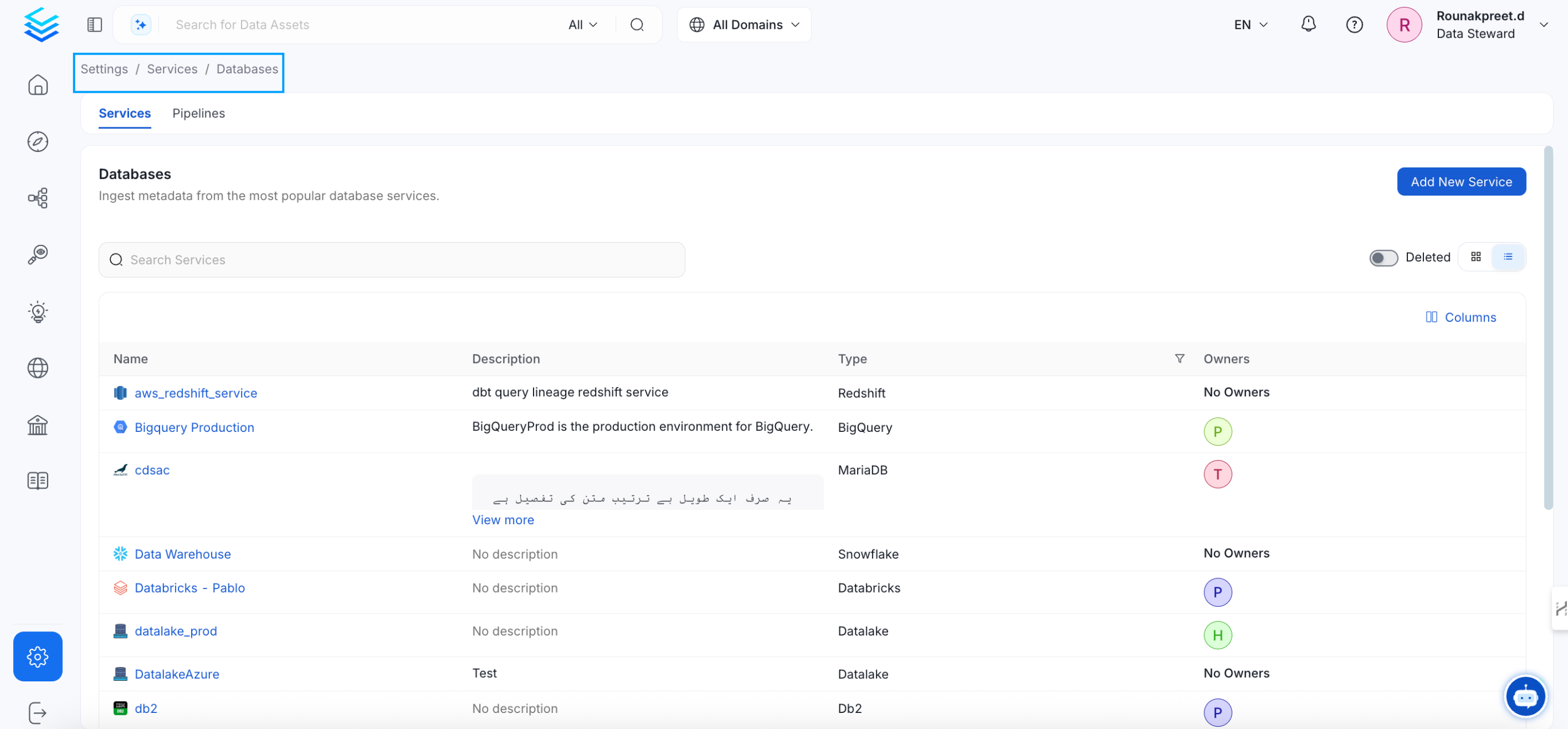
Add a new Service from the Services page

Select your Service from the list
4. Name and Describe your Service
Provide a name and description for your Service.
Service Name
OpenMetadata uniquely identifies Services by their Service Name. Provide a name that distinguishes your deployment from other Services, including the other BigQuery Services that you might be ingesting metadata from.
Note that when the name is set, it cannot be changed.
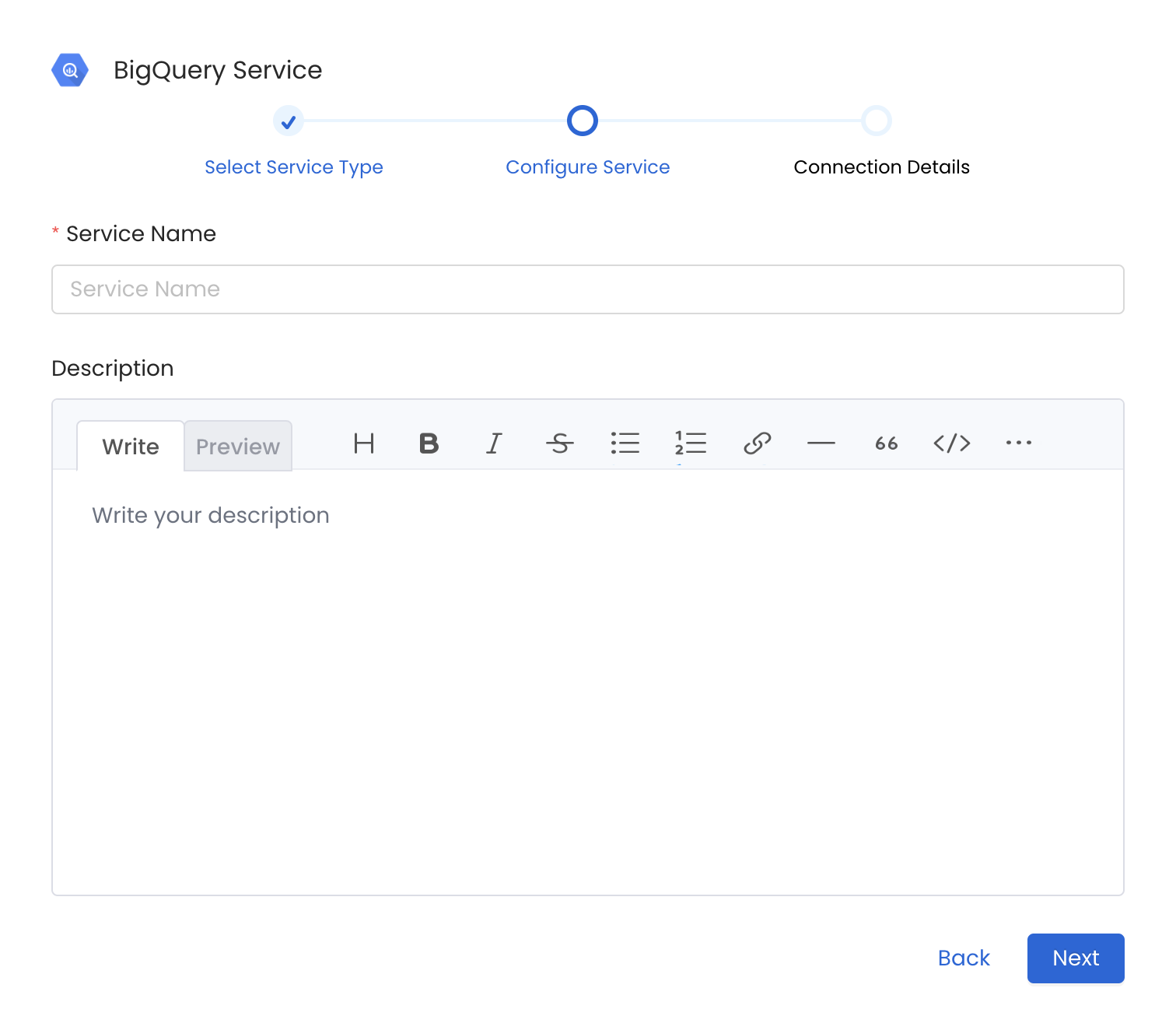
Provide a Name and description for your Service
5. Configure the Service Connection
In this step, we will configure the connection settings required for BigQuery.
Please follow the instructions below to properly configure the Service to read from your sources. You will also find helper documentation on the right-hand side panel in the UI.
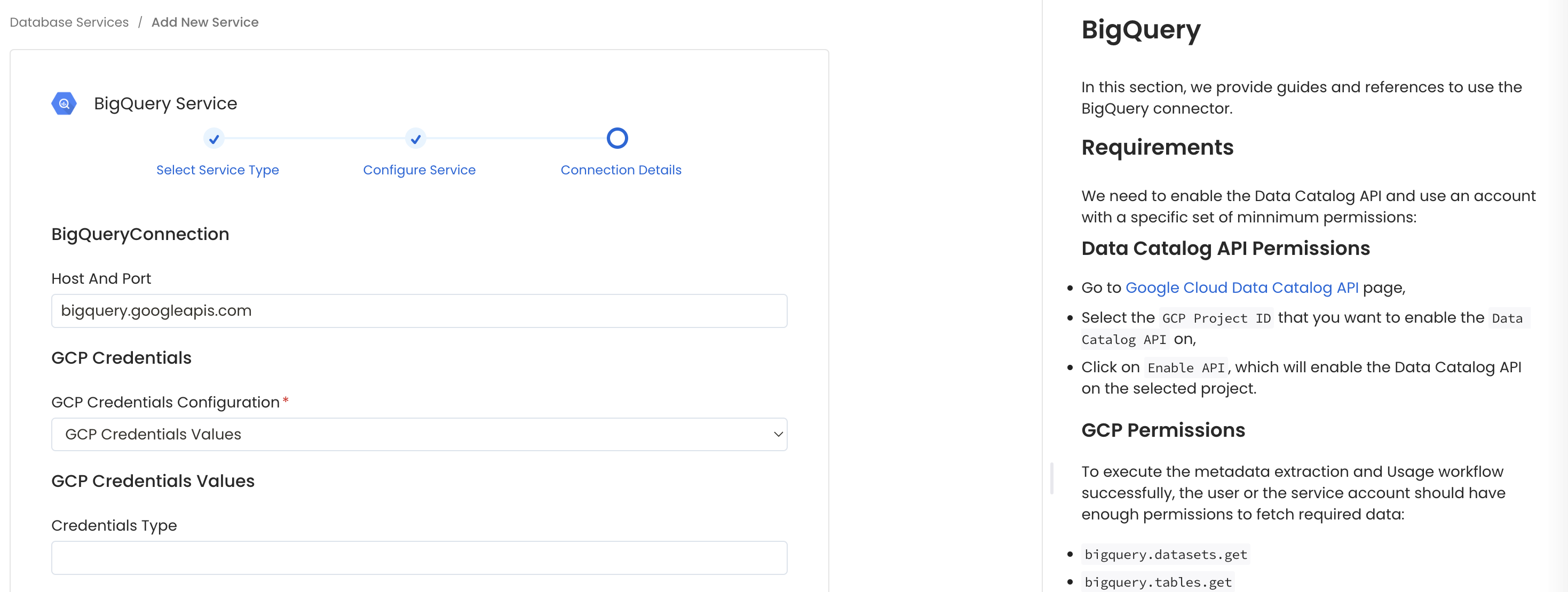
Configure the Service connection by filling the form
Connection Options
Host and Port: BigQuery APIs URL. By default, the API URL is bigquery.googleapis.com you can modify this if you have custom implementation of BigQuery.
GCP Credentials: You can authenticate with your bigquery instance using either GCP Credentials Path where you can specify the file path of the service account key or you can pass the values directly by choosing the GCP Credentials Values from the service account key file.
You can check out this documentation on how to create the service account keys and download it.
GCP Credentials Values: Passing the raw credential values provided by BigQuery. This requires us to provide the following information, all provided by BigQuery:
- Credentials type: Credentials Type is the type of the account, for a service account the value of this field is
service_account. To fetch this key, look for the value associated with thetypekey in the service account key file. - Billing Project ID (Optional): A billing project ID is a unique string used to identify and authorize your project for billing in Google Cloud.
- Project ID: A project ID is a unique string used to differentiate your project from all others in Google Cloud. To fetch this key, look for the value associated with the
project_idkey in the service account key file. You can also pass multiple project id to ingest metadata from different BigQuery projects into one service. - Private Key ID: This is a unique identifier for the private key associated with the service account. To fetch this key, look for the value associated with the
private_key_idkey in the service account file. - Private Key: This is the private key associated with the service account that is used to authenticate and authorize access to BigQuery. To fetch this key, look for the value associated with the
private_keykey in the service account file. - Client Email: This is the email address associated with the service account. To fetch this key, look for the value associated with the
client_emailkey in the service account key file. - Client ID: This is a unique identifier for the service account. To fetch this key, look for the value associated with the
client_idkey in the service account key file. - Auth URI: This is the URI for the authorization server. To fetch this key, look for the value associated with the
auth_urikey in the service account key file. The default value to Auth URI is https://accounts.google.com/o/oauth2/auth. - Token URI: The Google Cloud Token URI is a specific endpoint used to obtain an OAuth 2.0 access token from the Google Cloud IAM service. This token allows you to authenticate and access various Google Cloud resources and APIs that require authorization. To fetch this key, look for the value associated with the
token_urikey in the service account credentials file. Default Value to Token URI is https://oauth2.googleapis.com/token. - Authentication Provider X509 Certificate URL: This is the URL of the certificate that verifies the authenticity of the authorization server. To fetch this key, look for the value associated with the
auth_provider_x509_cert_urlkey in the service account key file. The Default value for Auth Provider X509Cert URL is https://www.googleapis.com/oauth2/v1/certs - Client X509Cert URL: This is the URL of the certificate that verifies the authenticity of the service account. To fetch this key, look for the value associated with the
client_x509_cert_urlkey in the service account key file.
GCP Credentials Path: Passing a local file path that contains the credentials.
Include Policy Tags (Optional): Enable this to ingest BigQuery policy tags. Make sure the Include Tags option is enabled in the ingestion agent. If Include Policy Tags is disabled, the agent will only ingest labels according to the Include Tags setting.
Taxonomy Project ID (Optional): Bigquery uses taxonomies to create hierarchical groups of policy tags. To apply access controls to BigQuery columns, tag the columns with policy tags. Learn more about how yo can create policy tags and set up column-level access control here
If you have attached policy tags to the columns of table available in Bigquery, then OpenMetadata will fetch those tags and attach it to the respective columns.
In this field you need to specify the id of project in which the taxonomy was created.
Taxonomy Location (Optional): Bigquery uses taxonomies to create hierarchical groups of policy tags. To apply access controls to BigQuery columns, tag the columns with policy tags. Learn more about how yo can create policy tags and set up column-level access control here
If you have attached policy tags to the columns of table available in Bigquery, then OpenMetadata will fetch those tags and attach it to the respective columns.
In this field you need to specify the location/region in which the taxonomy was created.
Usage Location (Optional): Location used to query INFORMATION_SCHEMA.JOBS_BY_PROJECT to fetch usage data. You can pass multi-regions, such as us or eu, or your specific region such as us-east1. Australia and Asia multi-regions are not yet supported.
Cost Per TiB (Optional): The cost (in USD) per tebibyte (TiB) of data processed during BigQuery usage analysis. This value is used to estimate query costs when analyzing usage metrics from INFORMATION_SCHEMA.JOBS_BY_PROJECT.
This setting does not affect actual billing—it is only used for internal reporting and visualization of estimated costs.
The default value, if not set, may assume the standard on-demand BigQuery pricing (e.g., $5.00 per TiB), but you should adjust it according to your organization's negotiated rates or flat-rate pricing model.
Application Default Credentials (ADC) Authentication
If you want to use ADC authentication for BigQuery, configure the GCP credentials with type gcp_adc:
Using ADC with Billing Project ID: When using ADC authentication, you can still specify a Billing Project ID to ensure proper billing attribution for your BigQuery queries. This is particularly useful when:
- Your service account has access to multiple projects
- You want to bill queries to a specific project different from the one containing your data
- You're running queries that span multiple projects
ADC Setup: ADC authentication works automatically when running in Google Cloud environments (GKE, Compute Engine, Cloud Run) or when you've configured it locally using gcloud auth application-default login.
Advanced Configuration
Database Services have an Advanced Configuration section, where you can pass extra arguments to the connector and, if needed, change the connection Scheme.
This would only be required to handle advanced connectivity scenarios or customizations.
- Connection Options (Optional): Enter the details for any additional connection options that can be sent to database during the connection. These details must be added as Key-Value pairs.
- Connection Arguments (Optional): Enter the details for any additional connection arguments such as security or protocol configs that can be sent during the connection. These details must be added as Key-Value pairs.
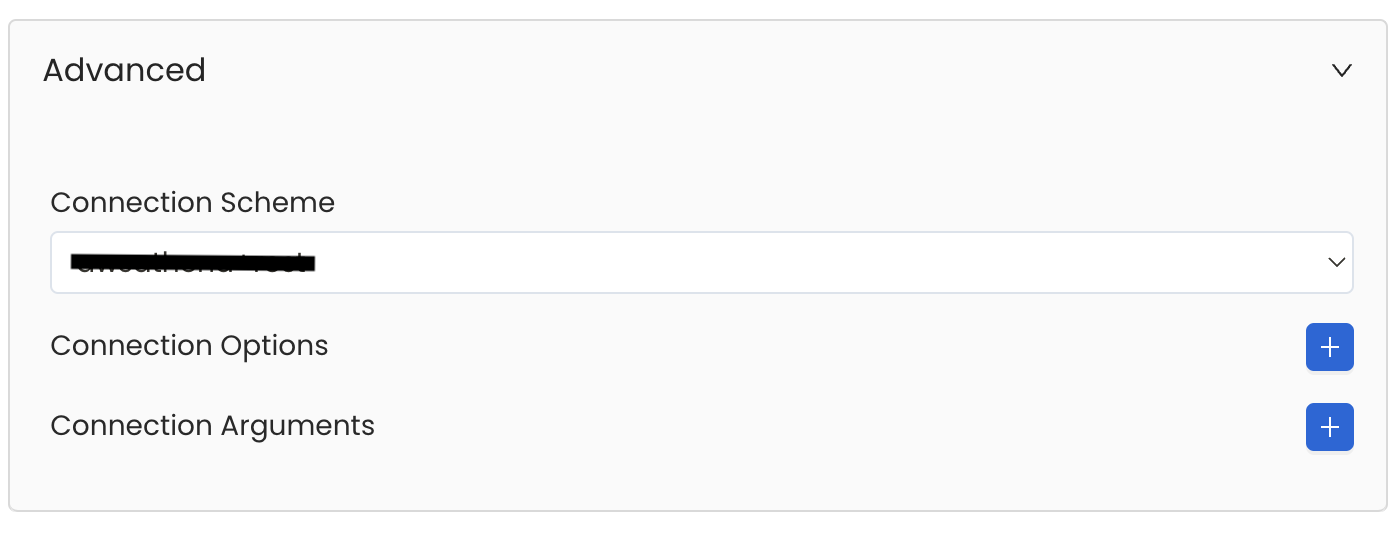
Advanced Configuration
6. Test the Connection
Once the credentials have been added, click on Test Connection and Save the changes.
Test the connection and save the Service
7. Configure Metadata Ingestion
In this step we will configure the metadata ingestion pipeline, Please follow the instructions below
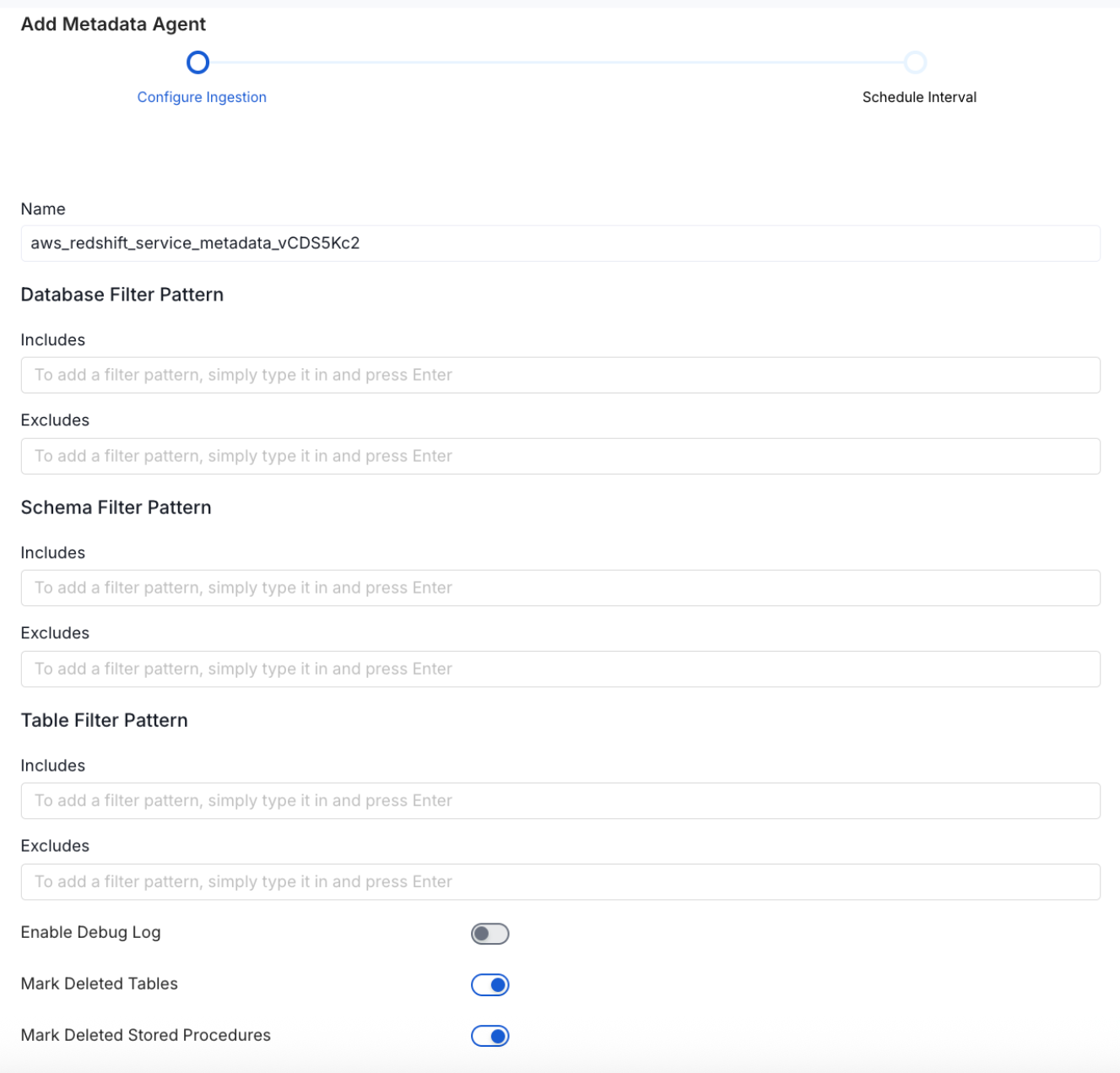
Configure Metadata Ingestion Page - 1
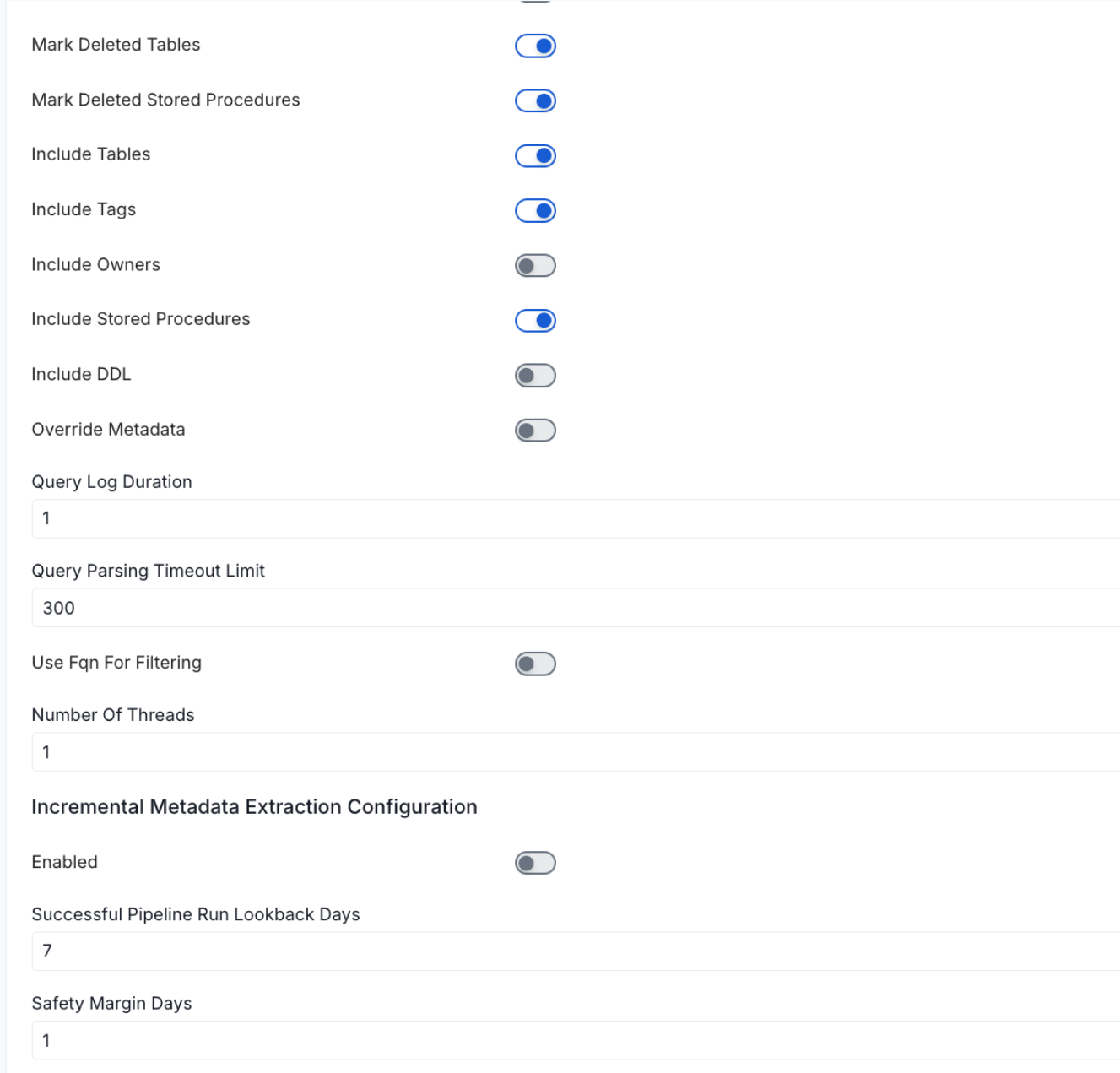
Configure Metadata Ingestion Page - 2
Metadata Ingestion Options
If the owner's name is openmetadata, you need to enter openmetadata@domain.com in the name section of add team/user form, click here for more info.
Name: This field refers to the name of ingestion pipeline, you can customize the name or use the generated name.
Database Filter Pattern (Optional): Use to database filter patterns to control whether or not to include database as part of metadata ingestion.
- Include: Explicitly include databases by adding a list of comma-separated regular expressions to the Include field. OpenMetadata will include all databases with names matching one or more of the supplied regular expressions. All other databases will be excluded.
- Exclude: Explicitly exclude databases by adding a list of comma-separated regular expressions to the Exclude field. OpenMetadata will exclude all databases with names matching one or more of the supplied regular expressions. All other databases will be included.
Schema Filter Pattern (Optional): Use to schema filter patterns to control whether to include schemas as part of metadata ingestion.
- Include: Explicitly include schemas by adding a list of comma-separated regular expressions to the Include field. OpenMetadata will include all schemas with names matching one or more of the supplied regular expressions. All other schemas will be excluded.
- Exclude: Explicitly exclude schemas by adding a list of comma-separated regular expressions to the Exclude field. OpenMetadata will exclude all schemas with names matching one or more of the supplied regular expressions. All other schemas will be included.
Table Filter Pattern (Optional): Use to table filter patterns to control whether to include tables as part of metadata ingestion.
- Include: Explicitly include tables by adding a list of comma-separated regular expressions to the Include field. OpenMetadata will include all tables with names matching one or more of the supplied regular expressions. All other tables will be excluded.
- Exclude: Explicitly exclude tables by adding a list of comma-separated regular expressions to the Exclude field. OpenMetadata will exclude all tables with names matching one or more of the supplied regular expressions. All other tables will be included.
Enable Debug Log (toggle): Set the Enable Debug Log toggle to set the default log level to debug.
Mark Deleted Tables (toggle): Set the Mark Deleted Tables toggle to flag tables as soft-deleted if they are not present anymore in the source system.
Mark Deleted Tables from Filter Only (toggle): Set the Mark Deleted Tables from Filter Only toggle to flag tables as soft-deleted if they are not present anymore within the filtered schema or database only. This flag is useful when you have more than one ingestion pipelines. For example if you have a schema
includeTables (toggle): Optional configuration to turn off fetching metadata for tables.
includeViews (toggle): Set the Include views toggle to control whether to include views as part of metadata ingestion.
includeTags (toggle): Set the 'Include Tags' toggle to control whether to include tags as part of metadata ingestion.
includeOwners (toggle): Set the 'Include Owners' toggle to control whether to include owners to the ingested entity if the owner email matches with a user stored in the OM server as part of metadata ingestion. If the ingested entity already exists and has an owner, the owner will not be overwritten.
includeStoredProcedures (toggle): Optional configuration to toggle the Stored Procedures ingestion.
includeDDL (toggle): Optional configuration to toggle the DDL Statements ingestion.
queryLogDuration (Optional): Configuration to tune how far we want to look back in query logs to process Stored Procedures results.
queryParsingTimeoutLimit (Optional): Configuration to set the timeout for parsing the query in seconds.
useFqnForFiltering (toggle): Regex will be applied on fully qualified name (e.g service_name.db_name.schema_name.table_name) instead of raw name (e.g. table_name).
Incremental (Beta): Use Incremental Metadata Extraction after the first execution. This is done by getting the changed tables instead of all of them. Only Available for BigQuery, Redshift and Snowflake
- Enabled: If
True, enables Metadata Extraction to be Incremental. - lookback Days: Number of days to search back for a successful pipeline run. The timestamp of the last found successful pipeline run will be used as a base to search for updated entities.
- Safety Margin Days: Number of days to add to the last successful pipeline run timestamp to search for updated entities.
- Enabled: If
Threads (Beta): Use a Multithread approach for Metadata Extraction. You can define here the number of threads you would like to run concurrently. For further information please check the documentation on Metadata Ingestion - Multithreading
Note that the right-hand side panel in the OpenMetadata UI will also share useful documentation when configuring the ingestion.
8. Schedule the Ingestion and Deploy
Scheduling can be set up at an hourly, daily, weekly, or manual cadence. The timezone is in UTC. Select a Start Date to schedule for ingestion. It is optional to add an End Date.
Review your configuration settings. If they match what you intended, click Deploy to create the service and schedule metadata ingestion.
If something doesn't look right, click the Back button to return to the appropriate step and change the settings as needed.
After configuring the workflow, you can click on Deploy to create the pipeline.
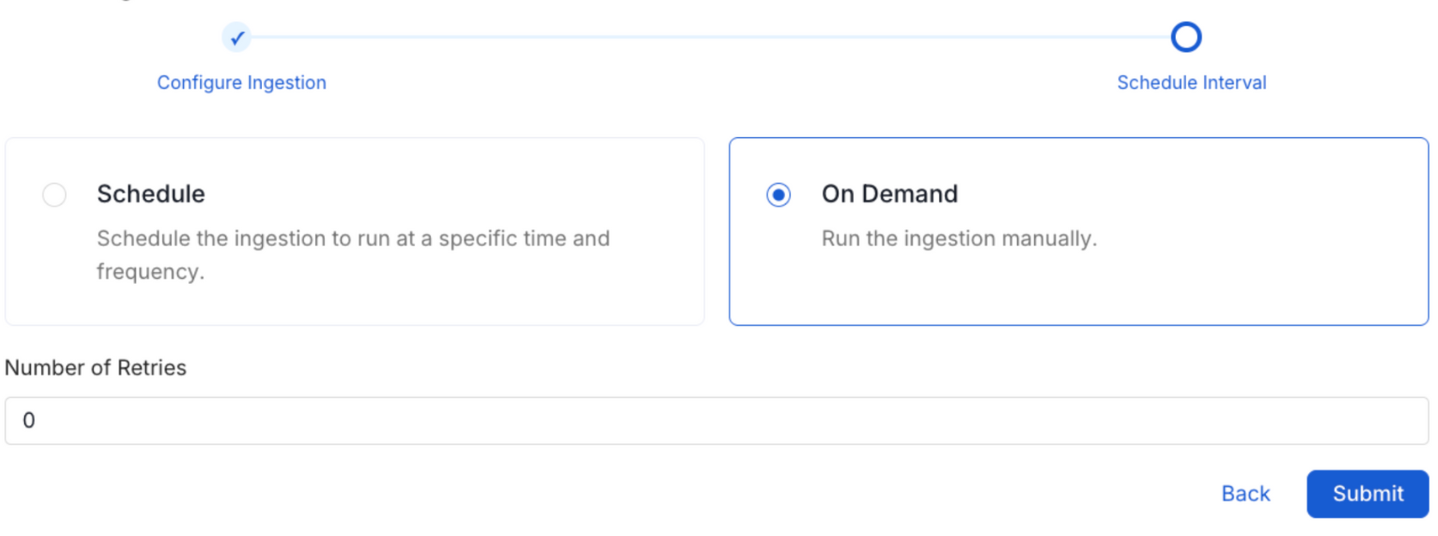
Schedule the Ingestion Pipeline and Deploy
Cross Project Lineage
We support cross-project lineage, but the data must be ingested within a single service. This means you need to perform lineage ingestion for just one service while including multiple projects.
